

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2025, Vol. 34 ›› Issue (11): 136-149.DOI: 10.11686/cyxb2024480
• 研究论文 • 上一篇
付瑶1,2( ), 王子贤1,2, 陈泰祥3, 晋玲1,2, 马晓辉1,2, 王艳1,2(
), 王子贤1,2, 陈泰祥3, 晋玲1,2, 马晓辉1,2, 王艳1,2( )
)
收稿日期:2024-12-03
修回日期:2025-03-03
出版日期:2025-11-20
发布日期:2025-10-09
通讯作者:
王艳
作者简介:E-mail: gswangyan101@163.com基金资助:
Yao FU1,2( ), Zi-xian WANG1,2, Tai-xiang CHEN3, Ling JIN1,2, Xiao-hui MA1,2, Yan WANG1,2(
), Zi-xian WANG1,2, Tai-xiang CHEN3, Ling JIN1,2, Xiao-hui MA1,2, Yan WANG1,2( )
)
Received:2024-12-03
Revised:2025-03-03
Online:2025-11-20
Published:2025-10-09
Contact:
Yan WANG
摘要:
党参黄芯病降低党参的产量和质量,在明确该病病原菌分类地位的基础上,构建增强型绿色荧光蛋白(enhanced green fluorescent protein, EGFP)标记的党参黄芯病菌转化株,为研究病原菌在党参植株体内的侵染特性提供可视化跟踪手段。基于形态学和多基因位点联合的方法,明确病原菌的分类地位。采用根癌农杆菌介导法(Agrobacteriumtumefaciens-mediated transformation, ATMT),将带有潮霉素磷酸转移酶基因(hygromycin phosphotransferase, HygB)和增强型绿色荧光蛋白基因EGFP的双元载体转入党参黄芯病菌分生孢子中并筛选出遗传稳定的转化子。结果表明,党参黄芯病常年田间发病率为20%~30%。病原菌为弯镰孢菌党参变种,遗传转化共获得138株阳性转化子,转化效率约为46个转化子·10-6个孢子。随机挑选的4株阳性转化子经6次继代培养,具有稳定的潮霉素B抗性和绿色荧光表达,其中转化子EGFP7-3与EGFP7-4生物学特性及致病性和野生型党参黄芯病菌无显著差异,表明绿色荧光蛋白基因已成功转入党参黄芯病菌中,转化子EGFP7-3与EGFP7-4稳定遗传且致病力不受影响。本研究为党参黄芯病菌致病机理的研究提供了良好的材料和技术支撑。
付瑶, 王子贤, 陈泰祥, 晋玲, 马晓辉, 王艳. 党参黄芯病菌的鉴定及绿色荧光蛋白基因转化[J]. 草业学报, 2025, 34(11): 136-149.
Yao FU, Zi-xian WANG, Tai-xiang CHEN, Ling JIN, Xiao-hui MA, Yan WANG. Identification and green fluorescent protein gene transformation of the causal agent of yellow-core disease of Codonopsis pilosula[J]. Acta Prataculturae Sinica, 2025, 34(11): 136-149.

图3 DSHX2显微形态A~C: PDA、CLA和SNA上菌落形态Colonies on PDA, CLA and SNA media; D: 大型分生孢子Macroconidia; E: 小型分生孢子Microconidia; F: 厚垣孢子Chlamydospores (标尺Scale bar=25 μm).
Fig.3 Morphology of DSHX2

图5 接种黄芯病病原菌后党参根系的发病症状A, E: 无伤接种Noninvasive inoculation; B, F: 刺伤接种Stab inoculation; C, G: 无伤对照Noninvasive blank control; D, H: 刺伤对照Stab control.
Fig.5 Symptoms in roots of C. pilosula after inoculation with yellow core disease pathogen
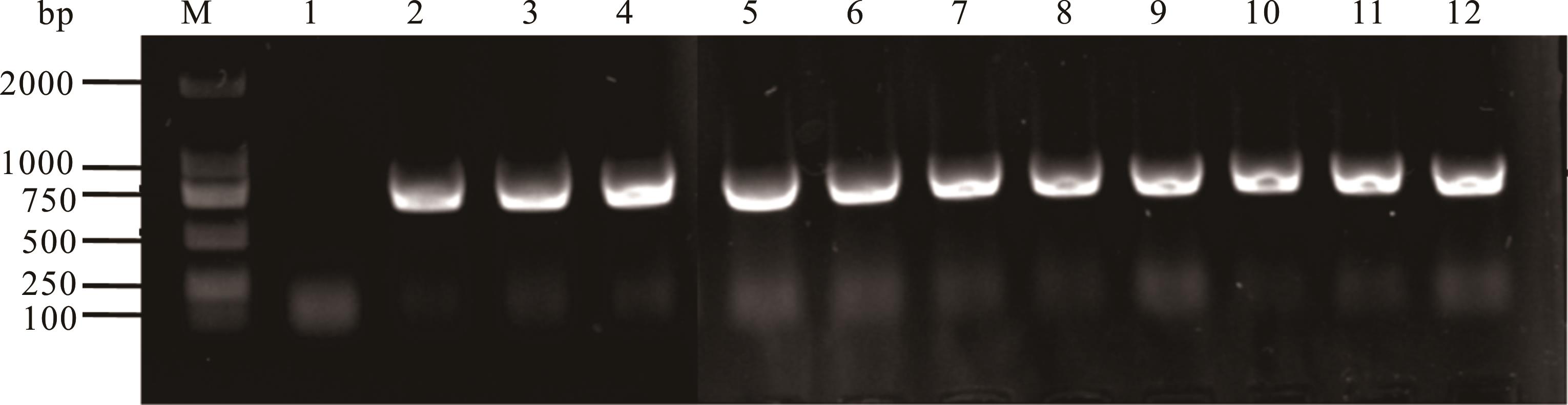
图6 阳性根癌农杆菌EGFP基因的PCR验证结果M: Marker; 1: 蒸馏水,阴性对照Sterile water, negative control; 2: pCAMBIA1303-gpdA-EGFP-TrpC-Hygro,阳性对照Positive control; 3~12: EGFP农杆菌阳性转化子Transformed A. tumefaciens strains harboring EGFP.
Fig.6 PCR verification results of the positive A. tumefaciens EGFP gene
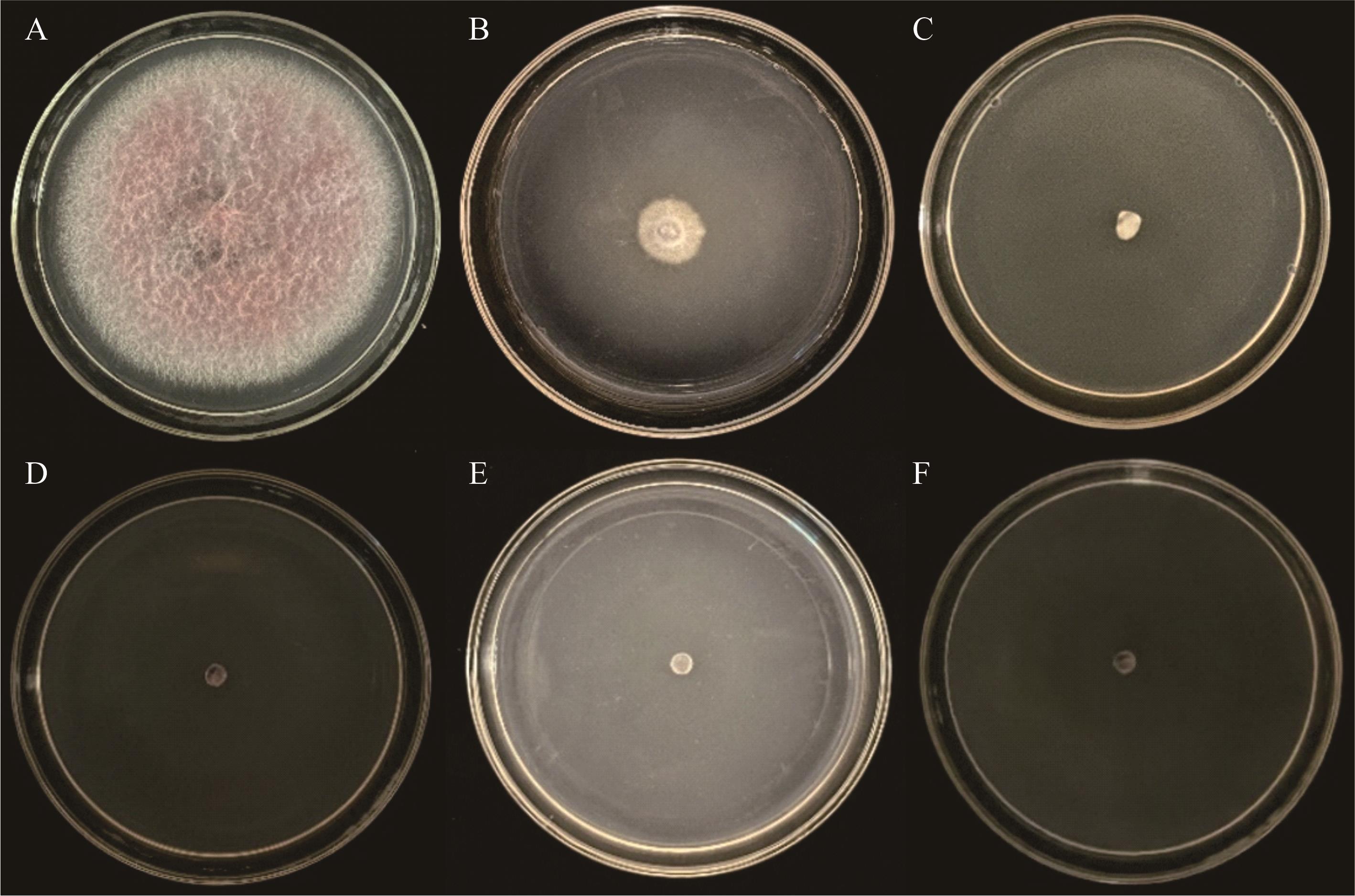
图7 不同浓度HygB对DSHX2的生长抑制情况A: 0 μg·mL-1 HygB; B: 25 μg·mL-1 HygB; C: 50 μg·mL-1 HygB; D: 100 μg·mL-1 HygB; E: 150 μg·mL-1 HygB; F: 200 μg·mL-1 HygB.
Fig.7 DSHX2 growth rate under different concentrations of hygromycin B
项目 Item | 头孢噻肟钠浓度Mass concentration of cefotaxime sodium | ||||
|---|---|---|---|---|---|
| 0 μg·mL-1 | 25 μg·mL-1 | 50 μg·mL-1 | 100 μg·mL-1 | 150 μg·mL-1 | |
| 农杆菌生长状况The growth state of LBA4404 | ++ | + | - | - | - |
表1 农杆菌生长受头孢噻肟钠的抑制作用
Table 1 Agrobacterium growth rate under different concentrations of cefotaxime sodium
项目 Item | 头孢噻肟钠浓度Mass concentration of cefotaxime sodium | ||||
|---|---|---|---|---|---|
| 0 μg·mL-1 | 25 μg·mL-1 | 50 μg·mL-1 | 100 μg·mL-1 | 150 μg·mL-1 | |
| 农杆菌生长状况The growth state of LBA4404 | ++ | + | - | - | - |
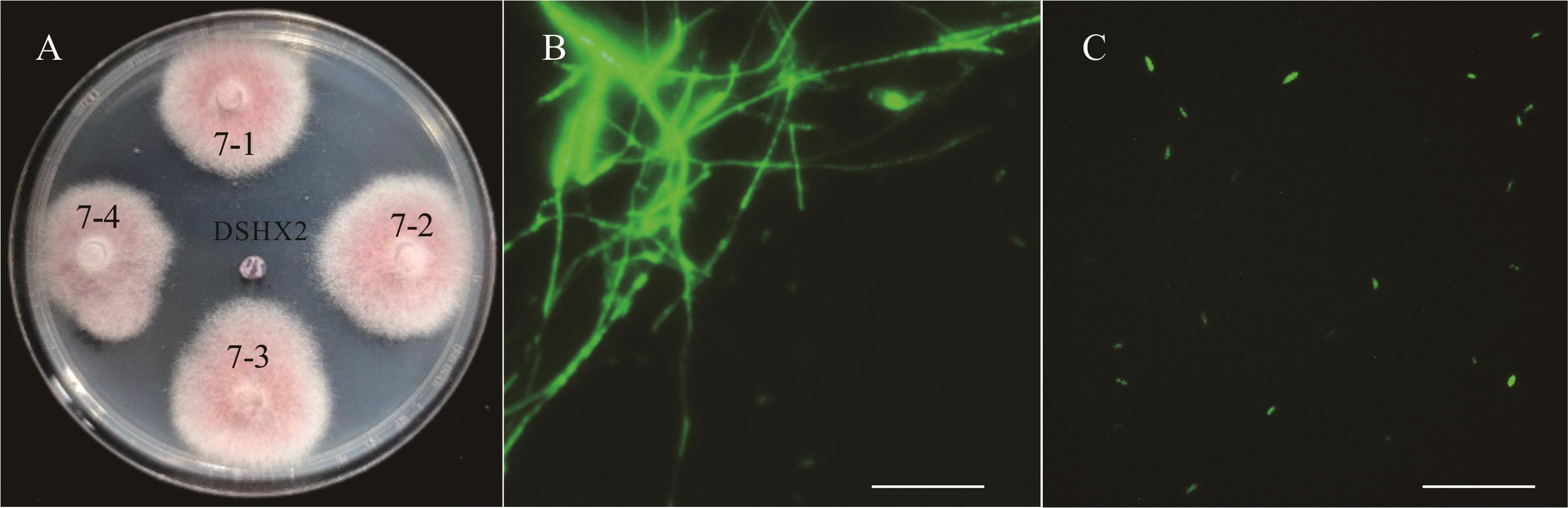
图8 阳性转化子HygB稳定性和增强型绿色荧光表达A为含有100 μg·mL-1 HygB的PDA平板上4株转化子和DSHX2 Four transformants and DSHX2 grown on PDA supplemented with 100 μg·mL-1 hygromycin;B, C分别为4号转化子(EGFP7-4)荧光显微镜蓝色激发光下菌丝和分生孢子Mycelia and conidia of EGFP7-4, respectively, under blue excitation light microscopy (标尺Scale bar=50 μm).
Fig.8 Stability of hygromycin-resistant transformants and enhanced green fluorescent protein expression
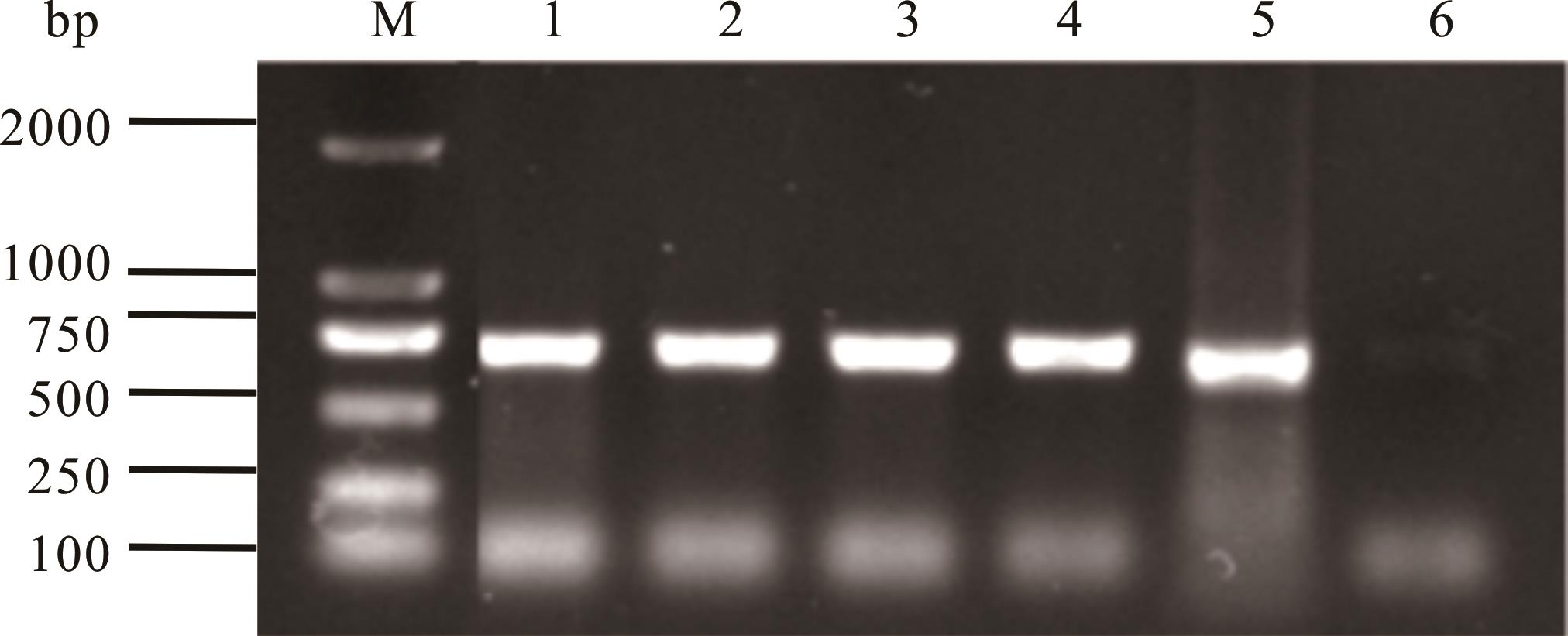
图9 转化子EGFP基因PCR验证M: DNA Marker; 1~4: 转化子EGFP7-1~EGFP7-4 Transformants EGFP7-1-EGFP7-4; 5: pCAMBIA1303-gpdA-EGFP-TrpC-Hygro,阳性对照Positive control; 6: DSHX2,阴性对照Negative control.
Fig.9 PCR verification of EGFP gene of the transformants
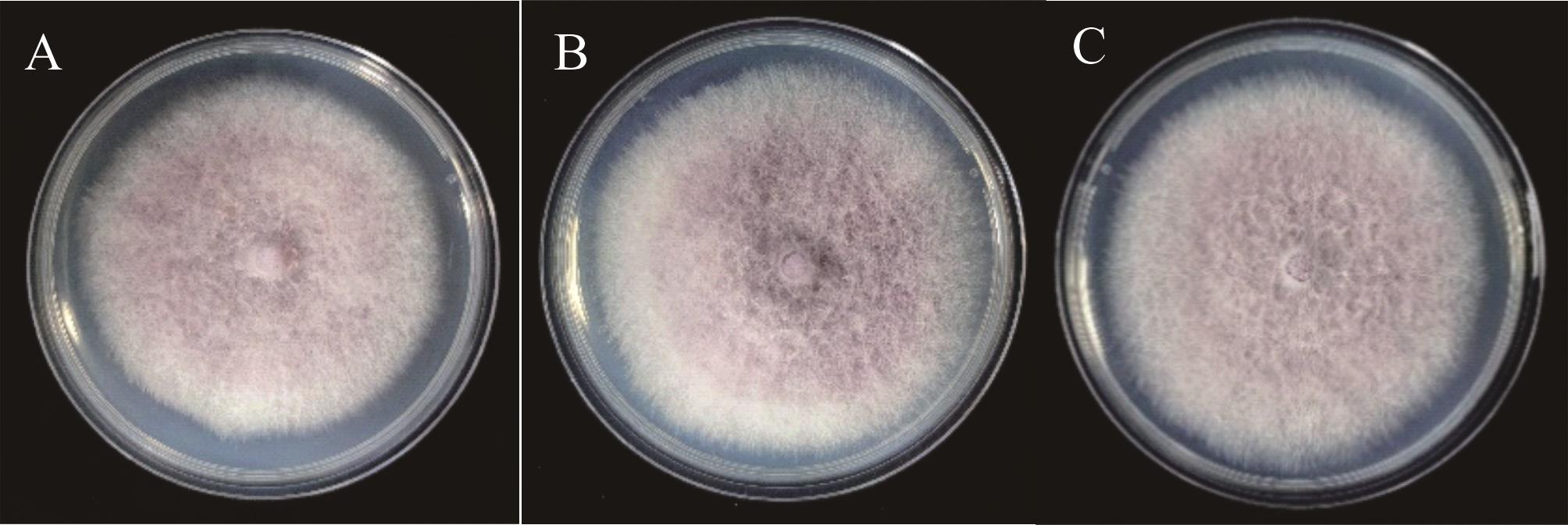
图10 DSHX2与阳性转化子的菌落形态A: DSHX2;野生型菌株The wild type strain; B, C: EGFP7-3、EGFP7-4,阳性转化子Positive transformants.
Fig.10 The colony morphology of DSHX2 and positive transformants of F. curvatum var. codonopsidis
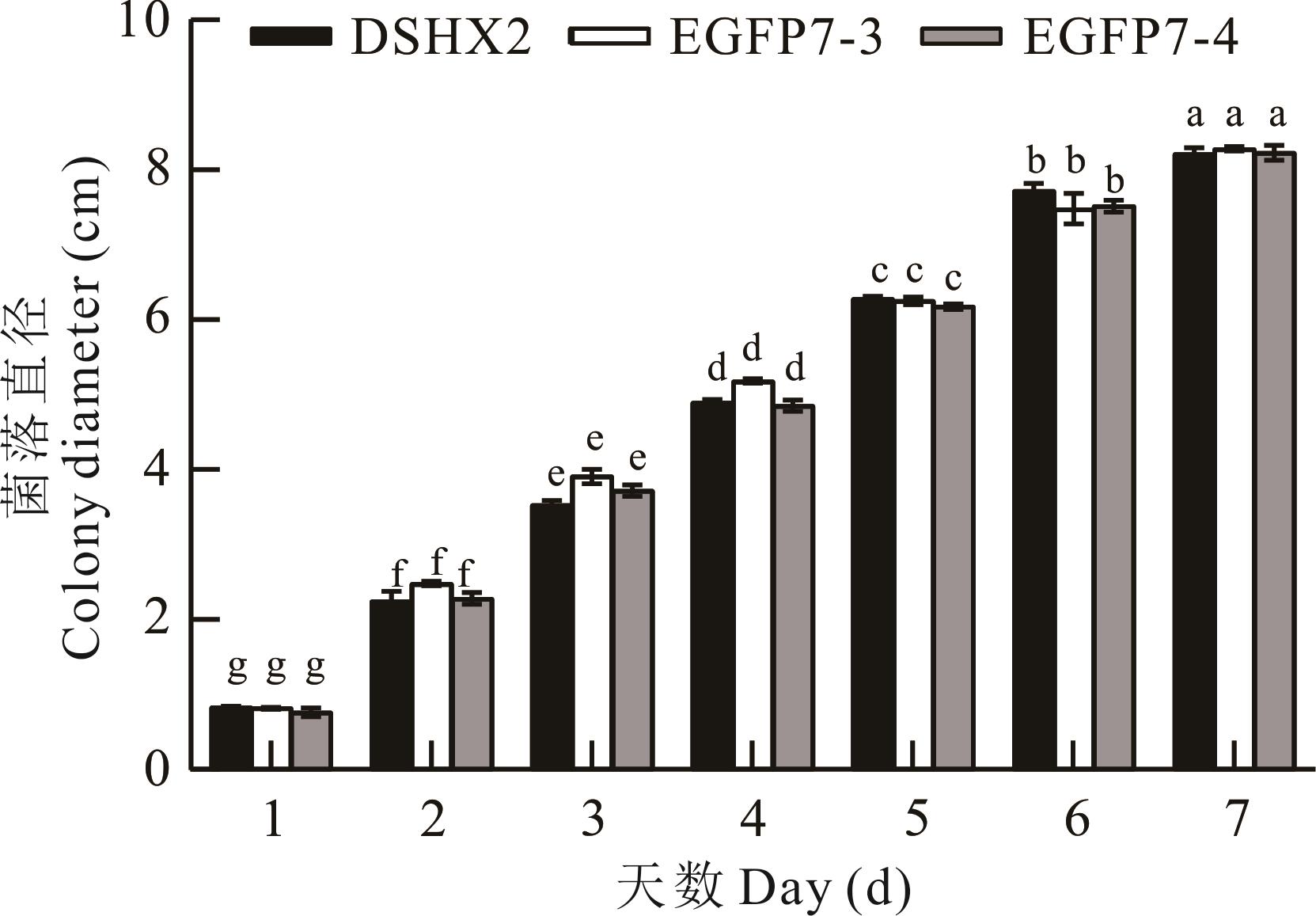
图11 野生子与转化子在PDA平板上的菌落直径不同小写字母表示差异显著(P<0.05)。Different lowercase letters mean significant difference at P<0.05 level.
Fig.11 Colony diameter of wild-type and positive transformants on PDA
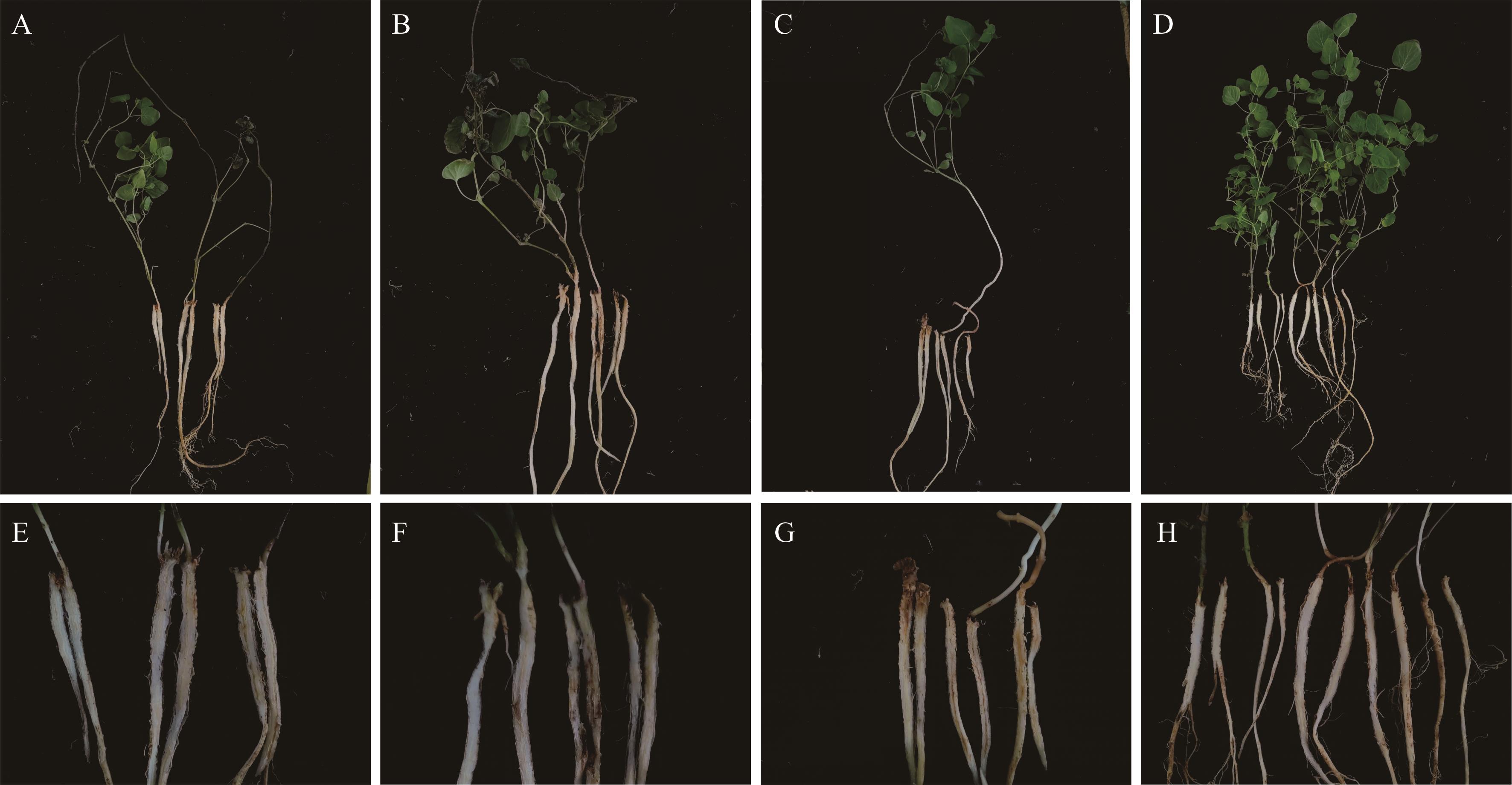
图13 转化子EGFP7-3和EGFP7-4与野生子DSHX2致病性测定A, E: EGFP7-3; B, F: EGFP7-4; C, G: DSHX2; D, H: 无菌水Sterile water.
Fig.13 Pathogenicity test of transformants EGFP7-3,EGFP7-4 and DSHX2
| [1] | State Pharmacopoeia Commission. People’s Republic of China Pharmacopoeia (one). Beijing: China Medical Science and Technology Press, 2020: 342. |
| 国家药典委员会. 中国药典(一部). 北京: 中国医药科技出版社, 2020: 342. | |
| [2] | Li F, Yang F D. Research progress on extraction and isolation, chemical compound and pharmacological effects of polysaccharide from Dangshen (Codonopsis Radix). Chinese Archives of Traditional Chinese Medicine, 2023, 41(4): 42-49. |
| 李芳, 杨扶德. 党参多糖提取分离、化学组成和药理作用研究进展. 中华中医药学刊, 2023, 41(4): 42-49. | |
| [3] | Li C, Zhang H, Peng L, et al. Study of volatile chemical components in Codonopsis pilosula. Journal of Yunnan University (Natural Sciences Edition), 1993, 15(S2): 86-90. |
| 李聪, 张红, 彭莉,等. 党参中挥发性化学成分的研究. 云南大学学报(自然科学版), 1993, 15(S2): 86-90. | |
| [4] | Chen J Y, Liu Z G, Liu Q L, et al. Ozone application suppressed the blue mold development and maintained the main active ingredients content of postharvest fresh Codonopsis pilosula during storage. Journal of Fungi, 2024, 10(3): 163. |
| [5] | Zhao X, Liang Y, Constantine U, et al. First report of root rot caused by the Fusarium oxysporum species complex on Codonopsis pilosula in China. Plant Disease, 2021, (2021-06-02)[2025-03-03].Doi: 10.1094/PDIS-02-21-0418-PDN. |
| [6] | Yu Z L, Lei M Y, Pu S C, et al. Fungal disease survey and pathogen identification on Codonopsis tangshen in Chongqing. Journal of Chinese Medicinal Materials, 2015, 38(6): 1119-1122. |
| 余中莲, 雷美艳, 蒲盛才, 等. 重庆党参真菌病害种类调查及病原鉴定. 中药材, 2015, 38(6): 1119-1122. | |
| [7] | Chen S Z. Survey and control on grey mould in Codonopsis pilosula in Dingxi of Gansu Province. Grassland and Turf, 2017, 37(2): 94-97. |
| 陈书珍. 甘肃省定西党参灰霉病调查及田间药剂防治. 草原与草坪, 2017, 37(2): 94-97. | |
| [8] | Wang Y, Zeng C Y, Chen H G, et al. Pathogen identification and cultural characteristics of leaf spot of Codonopsis pilosula. Journal of Plant Protection, 2016, 43(6): 928-934. |
| 王艳, 曾翠云, 陈红刚,等. 党参斑枯病病原菌及其生物学特性. 植物保护学报, 2016, 43(6): 928-934. | |
| [9] | Sun X R, Zhong C P, Zhang X M, et al. Identification and control of the root rot pathogen of Codonopsis pilosula in Dingxi of Gansu Province. Plant Protection, 2020, 46(5): 290-297. |
| 孙新荣, 仲彩萍, 张西梅,等. 甘肃定西地区党参根腐病病原鉴定与防治研究. 植物保护, 2020, 46(5): 290-297. | |
| [10] | Wang Y, Chen X R, Wang Y Q, et al. Disease survey and pathogen identification on Codonopsis pilosula in Gansu Province. Journal of Shanxi Agricultural Sciences, 2011, 39(8): 866-868, 871. |
| 王艳, 陈秀蓉, 王引权,等. 甘肃省党参病害种类调查及病原鉴定. 山西农业科学, 2011, 39(8): 866-868, 871. | |
| [11] | Zou R, Ma J N, Zhang F. Research progress on Agrobacterium tumefaciens-mediated transformation system of mycorrhizal fungi. Journal of Fungal Research, 2021, 19(3): 197-206. |
| 邹嵘, 马嘉楠, 张凤. 根癌农杆菌介导的菌根真菌遗传转化研究进展. 菌物研究, 2021, 19(3): 197-206. | |
| [12] | Walter I, Fleischmann M, Klein D, et al. Rapid and sensitive detection of enhanced green fluorescent protein expression in paraffin sections by confocal laser scanning microscopy. The Histochemical Journal, 2000, 32(2): 99-103. |
| [13] | Jiang M, Lv J X, Huang Y L, et al. Applications of green fluorescent protein in plant pathology. Plant Protection, 2011, 37(5): 39-43. |
| 蒋明, 吕枷薪, 黄余磊,等. 绿色荧光蛋白在植物病理学研究中的应用. 植物保护, 2011, 37(5): 39-43. | |
| [14] | Yoon J H, Kim Y J, Kim S Y, et al. Agrobacterium tumefaciens-mediated transformation of the aquatic fungus Phialemonium inflatum FBCC-F1546. Journal of Fungi, 2023, 9(12): 1158. |
| [15] | Henares B M, Debler J W, Farfan-Caceres L M, et al. Agrobacterium tumefaciens-mediated transformation and expression of GFP in Ascochyta lentis to characterize ascochyta blight disease progression in lentil. PLoS One, 2019, 14(10): e0223419. |
| [16] | Sarrocco S, Falasvhi N, Vergara M, et al. Use of Fusarium oxysporum f. sp. dianthi transformed with marker genes to follow colonization of carnation roots. Journal of Plant Pathology, 2007, 89(1): 47-54. |
| [17] | Chen Y P, Liu X, Xiao R F, et al. Transformation of green fluorescent protein of Fusarium oxysporum isolated from diseased bitter gourd. Fujian Journal of Agricultural Sciences, 2020, 35(11): 1228-1233. |
| 陈燕萍, 刘欣, 肖荣凤, 等. 苦瓜枯萎病原菌的绿色荧光蛋白基因标记. 福建农业学报, 2020, 35(11): 1228-1233. | |
| [18] | Liu X Y, Luo Z P, Cui W K, et al. Histological observation of the infection process in Rehmannia glutinosa by Fusarium oxysporum labeled with green fluorescent protein (GFP) gene. Mycosystema, 2015, 34(6): 1092-1100. |
| 刘晓燕, 罗振鹏, 崔伟康,等. 绿色荧光蛋白标记下镰刀菌侵染地黄的组织学观察. 菌物学报, 2015, 34(6): 1092-1100. | |
| [19] | Liu Y, Gu A X, Zhu Q. Construction and analysis of mutant library of cotton Fusarium wilt. Microbiology China, 2022, 49(1): 125-138. |
| 刘叶, 顾爱星, 朱琦. 棉花枯萎病菌突变体库的构建及分析. 微生物学通报, 2022, 49(1): 125-138. | |
| [20] | Zhang X. Studies on genetic diversity and transformation of the green fluorescent protein gene in Fusarium oxysporum f. sp. cubense, causal agent of banana Fusarium wilt. Danzhou: South China Agricultural University, 2007. |
| 张欣. 香蕉枯萎病菌遗传多态性及绿色荧光蛋白基因转化的研究. 儋州: 华南热带农业大学, 2007. | |
| [21] | Zhang T, Wang Y, Jin L, et al. Biological characteristics and indoor toxicity test of main pathogens of Chinese angelica (Angelica sinensis) root rot. Journal of Microbiology, 2024, 44(2): 71-78. |
| 张婷, 王艳, 晋玲, 等. 当归根腐病主要病原菌生物学特性及室内毒力测定. 微生物学杂志, 2024, 44(2): 71-78. | |
| [22] | Donnell K O, Kistler H C, Cigelnik E, et al. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(5): 2044-2049. |
| [23] | Hofstetter V, Miadlikowska J, Kauff F, et al. Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: a case study of the Lecanoromycetes (Ascomycota). Molecular Phylogenetics and Evolution, 2007, 44(1): 412-426. |
| [24] | Reeb V, Lutzoni F, Roux C. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution, 2004, 32(3): 1036-1060. |
| [25] | Zhou X F, Zhao J, Zhao H, et al. A rapid identification method of recombinant Agrobacterium PCR reaction. Journal of Jilin Normal University (Natural Science Edition), 2015, 36(2): 109-111. |
| 周晓馥, 赵晶, 赵航, 等. 一种快速的重组农杆菌PCR鉴定方法. 吉林师范大学学报(自然科学版), 2015, 36(2): 109-111. | |
| [26] | Dong Y, Wang S T. Agrobacterium tumefaciens-mediated transformation method for Fusarium oxysporum. Methods in Molecular Biology, 2022, 2391: 63-73. |
| [27] | Luo Z Q, Cheng L, Zhang X, et al. A rapid method of preparing DNA template of filamentous fungi for PCR amplification. Biotechnology Bulletin, 2015, 31(9): 79-83. |
| 罗中钦, 程琳, 张茜,等. 丝状真菌PCR模板DNA的快速制备方法. 生物技术通报, 2015, 31(9): 79-83. | |
| [28] | Geiser D M, Al-Hatmi A M S, Aoki T, et al. Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology, 2021, 111(7): 1074-1079. |
| [29] | Nikitin D A, Ivanova E A, Semenov M V, et al. Diversity, ecological characteristics and identification of some problematic phytopathogenic Fusarium in soil: a review. Diversity, 2023, 15(1): 49. |
| [30] | Akgül D S, Önder S, Savaş N G, et al. Molecular identification and pathogenicity of Fusarium species associated with wood canker, root and basal rot in turkish grapevine nurseries. Journal of Fungi, 2024, 10(7): 444. |
| [31] | Fang D Z, Liu X L, Chen X R, et al. Fusarium species and Fusarium oxysporum species complex genotypes associated with yam wilt in south-central China. Frontiers in Microbiology, 2020, 17(11): 1964. |
| [32] | Mirghasempour S A, Michailides T, Chen W, et al. Fusarium spp. associated with dendrobium officinale dieback disease in China. Journal of Fungi, 2022, 8(9): 919 . |
| [33] | Zhou Y Y, Zhang W, Li X H, et al. Fusarium species associated with cherry leaf spot in China. Plants, 2022, 11(20): 2760. |
| [34] | Li C H, Li X G, Sun W B, et al. Identification of Fusarium cugenangense as a causal agent of wilt disease on Pyrus pyrifolia in China. Journal of Integrative Agriculture, 2024, (2024-02-29)[2025-03-03].Doi: 10.1016/j.jia.2024.02.018. |
| [35] | Lombard L, Sandoval-Denis M, Lamprecht S C, et al. Epitypification of Fusarium oxysporum-clearing the taxonomic chaos. Persoonia, 2019, 43: 1-47. |
| [36] | Turland N J, Wiersema J H, Barrie F R, et al. International code of nomenclature for algae, fungi, and plants (Shenzhen code). Glashütten, Germany: Koeltz Botanical Books, 2018. |
| [37] | Chakraborty B N, Patterson N A, Kapoor M. An electroporation-based system for high-efficiency transformation of germinated conidia of filamentous fungi. Canadian Journal of Microbiology, 1991, 37(11): 858-863. |
| [38] | DeGroot M J, Bundock P, Hooykaas P J, et al. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nature Biotechnology, 1998, 16(9): 839-842. |
| [39] | Li D D, Tang Y, Lin J, et al. Methods for genetic transformation of filamentous fungi. Microbial Cell Factories, 2017, 16(1): 168. |
| [40] | Song C L, Li L J, Wen J Z. Construction of vector for genetic transformation of Phytophthora sojae. Journal of Northeast Agricultural University, 2009, 40(1): 9-12. |
| 宋传玲, 李丽珺, 文景芝. 大豆疫霉菌(Phytophthora sojae)增强型绿色荧光蛋白遗传转化载体的构建. 东北农业大学学报, 2009, 40(1): 9-12. | |
| [41] | Deng D J, Meng Y N, Deng E J, et al. Application of genetic selection system in filamentous fungi: a review. Microbiology China, 2019, 46(5): 1165-1178. |
| 邓大杰, 孟亚南, 邓二杰,等. 丝状真菌遗传筛选系统的研究及其应用. 微生物学通报, 2019, 46(5): 1165-1178. | |
| [42] | Ren J J, Wang L X, Gao H B, et al. Agrobacterium tumefaciens-mediated transformation of Fusarium oxyporum f. sp. niveum. Plant Protection, 2015, 41(1): 93-97. |
| 任俊杰, 王丽霞, 高洪波,等. 农杆菌介导的西瓜枯萎病菌遗传转化. 植物保护, 2015, 41(1): 93-97. | |
| [43] | Liu Y, Ayibaota·Tuohedabai, Guo N N, et al. Optimization of Agrobacterium tumefaciens-mediated transformation system for Fusarium oxysporum. Microbiology China, 2021, 48(9): 2991-3001. |
| 刘叶, 阿依保他·托合达白, 郭楠楠,等. 农杆菌介导的棉花枯萎病菌转化体系的优化. 微生物学通报, 2021, 48(9): 2991-3001. | |
| [44] | Dong Y H, Wang S T. Fusarium wilt: Methods and protocols. New York: Springer US, 2022: 63-73. |
| [45] | Liu Y Q, Li C, Xu X D, et al. Marking of Fusarium oxysporum f. sp. cubense with green fluorescent protein (GFP) and measuement of its stability. Journal of Jilin Agricultural University, 2013, 35(1): 15-18. |
| 刘勇勤, 李赤, 徐秀德,等. 利用绿色荧光蛋白(GFP)标记香蕉枯萎病菌及其稳定性检测. 吉林农业大学学报, 2013, 35(1): 15-18. | |
| [46] | Fan L, Zhang J, Yang L, et al. Agrobacterium tumefaciens-mediated transformation of Botrytis cinerea strain RoseBc-3. Journal of Huazhong Agricultural University, 2013, 32(2): 30-35. |
| 范雷, 张静, 杨龙,等. 农杆菌介导灰葡萄孢菌株RoseBc-3的遗传转化. 华中农业大学学报, 2013, 32(2): 30-35. | |
| [47] | Yao J A, Zhang H, Huang P, et al. Green fluorescent protein genetic marker of Fusarium oxysporum F-02 of stem rot disease on Cymbidium ensifolium. Fujian Journal of Agricultural Sciences, 2019, 34(1): 70-75. |
| 姚锦爱, 张鸿, 黄鹏,等. 建兰茎腐病原菌尖孢镰刀菌F-02的绿色荧光蛋白基因标记. 福建农业学报, 2019, 34(1): 70-75. | |
| [48] | Zhang Y, Zhang J, Gao J, et al. The colonization process of sunflower by a green fluorescent protein-tagged isolate of Verticillium dahliae and its seed transmission. Plant Disease, 2018, 102(9): 1772-1778. |
| [49] | García-Plazaola J I, Fernández-Marín B, Duke S O, et al. Autofluorescence: biological functions and technical applications. Plant Science, 2015, 236: 136-145. |
| [50] | Dumur T, Duncan S, Graumann K, et al. Probing the 3D architecture of the plant nucleus with microscopy approaches: challenges and solutions. Nucleus, 2019, 10(1): 181-212. |
| [51] | Kodama Y. Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS One, 2016, 11(3): Doi: 10.1371/journal.pone.0152484. |
| [52] | Jiang X, Pees T, Reinhold-Hurek B. Deep-learning-based removal of autofluorescence and fluorescence quantification in plant-colonizing bacteria in vivo. New Phytologist, 2022, 235(6): 2481-2495. |
| [1] | 蒋晶晶, 杜蕙, 陈爱昌, 李雪萍, 李敏权, 漆永红. 甘肃省党参菌核病病原菌鉴定及其生物学特性研究[J]. 草业学报, 2022, 31(12): 181-190. |
| [2] | 王琼, 段廷玉, 南志标. 箭筈豌豆炭疽病病原菌分离鉴定[J]. 草业学报, 2020, 29(6): 127-136. |
| [3] | 靳鹏博, 胡佳栋, 毛歌, 张志伟, 马存德, 梁宗锁, 董娟娥. 栽培密度对党参产量和次生代谢物含量的影响[J]. 草业学报, 2018, 27(3): 164-172. |
| [4] | 潘龙其, 张丽, 杨成德, 袁庆华, 王瑜, 苗丽宏. 紫花苜蓿根腐病原菌——拟枝孢镰刀菌的鉴定及其生物学特性研究[J]. 草业学报, 2015, 24(10): 88-98. |
| [5] | 赵亚兰, 陈垣, 郭凤霞, 郭志军, 刘希全. 冬播和春播育苗对党参苗栽产量和质量的影响[J]. 草业学报, 2015, 24(10): 139-148. |
| [6] | 卞静,陈泰祥,陈秀蓉,王涵琦,杨小利,王艳. 当归新病害——炭疽病病原鉴定及发病规律研究[J]. 草业学报, 2014, 23(6): 266-273. |
| [7] | 漆永红,徐冬,张彦梅,曹素芳,郑果,杨发荣,李敏权. 嘉峪关市洋葱基盘腐烂病病原鉴定及药剂室内毒力测定[J]. 草业学报, 2013, 22(5): 339-344. |
| [8] | 安惠惠,马晖玲,李坚,白生军,马祥. 农杆菌介导的Lyz-GFP基因对匍匐翦股颖Penn A-1转化和表达的研究[J]. 草业学报, 2012, 21(2): 141-148. |
| [9] | 古丽君,徐秉良,梁巧兰,薛应钰. 兰州市草坪禾草根腐病的发生及病原菌鉴定[J]. 草业学报, 2009, 18(4): 175-180. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||