

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2023, Vol. 32 ›› Issue (4): 112-128.DOI: 10.11686/cyxb2022139
• 研究论文 • 上一篇
尚盼盼, 曾兵, 屈明好, 李明阳, 杨兴云, 郑玉倩, 沈秉娜, 毕磊, 杨成, 曾兵( )
)
收稿日期:2022-03-30
修回日期:2022-05-13
出版日期:2023-04-20
发布日期:2023-01-29
通讯作者:
曾兵
作者简介:E-mail:zbin78@163.com基金资助:
Pan-pan SHANG, Bing ZENG, Ming-hao QU, Ming-yang LI, Xing-yun YANG, Yu-qian ZHENG, Bing-na SHEN, Lei BI, Cheng YANG, Bing ZENG( )
)
Received:2022-03-30
Revised:2022-05-13
Online:2023-04-20
Published:2023-01-29
Contact:
Bing ZENG
摘要:
淹水胁迫是影响植物生长发育和分布的重要非生物胁迫,对植物淹水胁迫的研究是解决近年来极端强降水天气下植物生产管理的关键。红三叶作为优质豆科牧草,耐淹性较差,长期水淹会导致烂根死亡。为研究红三叶淹水胁迫下的分子响应机理,本研究通过Illumina高通量测序平台,以耐涝型品种“红龙”淹水胁迫下0、8 和24 h 三个时间点的幼苗叶片为材料进行转录组测序,将测序数据与参考基因组比对后进行差异表达基因(DEGs)分析和功能注释。结果显示,与对照0 h相比,“红龙”在淹水胁迫8 h后,有5065个DEGs,其中,上调表达基因2442个,下调表达基因2623个;在淹水胁迫24 h后,有9022个DEGs,其中,上调表达基因4279个,下调表达基因4743个。基因本体数据库富集结果显示,DEGs 主要富集于代谢过程、细胞过程、生物调节、细胞、催化活性等条目;东京基因与基因组数据库富集结果显示, DEGs显著富集于植物激素信号调节、植物-病原互作、碳代谢和乙醛酸及二羧酸代谢等通路中,其中乙醛酸及二羧酸代谢通路中过氧化氢酶和甲酸脱氢酶等抗氧化酶相关基因高表达;并且发现差异表达的AP2/ERF、WRKY、bHLH、NAC、bZIP等重要转录因子在红三叶响应淹水胁迫中也发挥重要作用。最后利用qRT-PCR对DEGs进行表达模式的分析验证,发现其与RNA-Seq结果一致,证实了测序结果的准确性。本研究根据转录组信息对DEGs展开功能注释、代谢通路、转录因子等方面的分析研究,初步了解红三叶对淹水胁迫的分子响应机理,为后续候选基因功能挖掘提供了基础数据和理论支撑。
尚盼盼, 曾兵, 屈明好, 李明阳, 杨兴云, 郑玉倩, 沈秉娜, 毕磊, 杨成, 曾兵. 红三叶响应淹水胁迫的相关通路及差异表达基因分析[J]. 草业学报, 2023, 32(4): 112-128.
Pan-pan SHANG, Bing ZENG, Ming-hao QU, Ming-yang LI, Xing-yun YANG, Yu-qian ZHENG, Bing-na SHEN, Lei BI, Cheng YANG, Bing ZENG. Analysis of metabolic pathways and differentially expressed genes of Trifolium pratense responding to waterlogging stress[J]. Acta Prataculturae Sinica, 2023, 32(4): 112-128.
| 名称Primer | 正向引物 Forward primers (5′-3′) | 反向引物Reverse primers (5′-3′) |
|---|---|---|
| gene623 | GGGCAGAGTCCGTGGTATGATAATC | AAAGATCGCTGGGTTGCTCGTG |
| gene26262 | CAACAACAAACACCACCACCAACG | TTGAGTTACGACGGAGCAACACTG |
| gene11075 | TCTCTACGGTATGATCAACGATGCA | CCCCCATGGTCTTCTCCTAACA |
| gene44440 | CACCACAACCACAATCACAACCATC | GCTCCGCTACAACGTGCTAGTG |
| gene21760 | CCCCAACTTGAGTCAGAAGCATC | TCCCTTTGATGTGCTGCACC |
| gene21720 | CGGATGAGTTAGGCGGATGGTTG | CGACATCTCCTCTTCTCCCAACATC |
| gene21305 | TCATCCCCTAAGAGGCCATACAGAG | CTGCTTCAGCGGTATCAAATGTTCC |
| gene3571 | TGACTTGCTCAACAATCCAGACCTC | GGCATGATGGCTTCGGTGACTG |
| GAPDH | TCTGACCGTTAGACTTGAGAAGG | CTTGAGCTTACCCTCAGACTCCT |
表1 差异表达基因引物序列
Table 1 Primers of differentially expressed genes
| 名称Primer | 正向引物 Forward primers (5′-3′) | 反向引物Reverse primers (5′-3′) |
|---|---|---|
| gene623 | GGGCAGAGTCCGTGGTATGATAATC | AAAGATCGCTGGGTTGCTCGTG |
| gene26262 | CAACAACAAACACCACCACCAACG | TTGAGTTACGACGGAGCAACACTG |
| gene11075 | TCTCTACGGTATGATCAACGATGCA | CCCCCATGGTCTTCTCCTAACA |
| gene44440 | CACCACAACCACAATCACAACCATC | GCTCCGCTACAACGTGCTAGTG |
| gene21760 | CCCCAACTTGAGTCAGAAGCATC | TCCCTTTGATGTGCTGCACC |
| gene21720 | CGGATGAGTTAGGCGGATGGTTG | CGACATCTCCTCTTCTCCCAACATC |
| gene21305 | TCATCCCCTAAGAGGCCATACAGAG | CTGCTTCAGCGGTATCAAATGTTCC |
| gene3571 | TGACTTGCTCAACAATCCAGACCTC | GGCATGATGGCTTCGGTGACTG |
| GAPDH | TCTGACCGTTAGACTTGAGAAGG | CTTGAGCTTACCCTCAGACTCCT |
| 样品Samples | 测序数据Clean reads | 测序下机数据Clean bases | GC含量GC content (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| H0-1 | 21397140 | 6395717310 | 41.76 | 97.95 | 93.95 |
| H0-2 | 21857534 | 6532928926 | 41.69 | 98.02 | 94.09 |
| H0-3 | 19569973 | 5851935352 | 41.57 | 97.98 | 93.94 |
| H8-1 | 23040082 | 6890947266 | 42.35 | 98.12 | 94.36 |
| H8-2 | 19333333 | 5775427470 | 43.44 | 98.06 | 94.31 |
| H8-3 | 26332328 | 7873279856 | 41.72 | 98.16 | 94.44 |
| H24-1 | 18119992 | 5414773508 | 42.65 | 98.21 | 94.65 |
| H24-2 | 39179129 | 11705420840 | 42.69 | 98.06 | 94.27 |
| H24-3 | 20547211 | 6143134312 | 41.70 | 98.10 | 94.30 |
表2 各样品测序数据统计
Table 2 Statistic of sequencing data of each sample
| 样品Samples | 测序数据Clean reads | 测序下机数据Clean bases | GC含量GC content (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| H0-1 | 21397140 | 6395717310 | 41.76 | 97.95 | 93.95 |
| H0-2 | 21857534 | 6532928926 | 41.69 | 98.02 | 94.09 |
| H0-3 | 19569973 | 5851935352 | 41.57 | 97.98 | 93.94 |
| H8-1 | 23040082 | 6890947266 | 42.35 | 98.12 | 94.36 |
| H8-2 | 19333333 | 5775427470 | 43.44 | 98.06 | 94.31 |
| H8-3 | 26332328 | 7873279856 | 41.72 | 98.16 | 94.44 |
| H24-1 | 18119992 | 5414773508 | 42.65 | 98.21 | 94.65 |
| H24-2 | 39179129 | 11705420840 | 42.69 | 98.06 | 94.27 |
| H24-3 | 20547211 | 6143134312 | 41.70 | 98.10 | 94.30 |
样品 Samples | 比对数据 Total reads | 比对上的数据 Mapped reads | 比对到唯一位置的数据 Unique mapped reads | 比对到多处位置的数据 Multiple mapped reads | 比对到正链的数据 Reads map to ‘+’ | 比对到负链的数据 Reads map to ‘-’ |
|---|---|---|---|---|---|---|
| H0-1 | 42794280 | 35486057 | 31690935 | 3795122 | 16073999 | 16721352 |
| H0-2 | 43715068 | 36362495 | 32008861 | 4353634 | 15860703 | 16888759 |
| H0-3 | 39139946 | 32347565 | 28590712 | 3756853 | 14374026 | 15045589 |
| H8-1 | 46080164 | 37362124 | 33004646 | 4357478 | 16597665 | 17394638 |
| H8-2 | 38666666 | 30216711 | 26296339 | 3920372 | 12726728 | 13856534 |
| H8-3 | 52664656 | 43972928 | 39050460 | 4922468 | 19761602 | 20580516 |
| H24-1 | 36239984 | 28099032 | 24677641 | 3421391 | 12290831 | 13074940 |
| H24-2 | 78358258 | 63574337 | 55516961 | 8057376 | 27856971 | 29552246 |
| H24-3 | 41094422 | 34030303 | 30035515 | 3994788 | 15052963 | 15928683 |
表3 样品测序数据与所选参考基因组的序列比对结果统计
Table 3 Sequence comparison results between sample sequencing data and the selected reference genome
样品 Samples | 比对数据 Total reads | 比对上的数据 Mapped reads | 比对到唯一位置的数据 Unique mapped reads | 比对到多处位置的数据 Multiple mapped reads | 比对到正链的数据 Reads map to ‘+’ | 比对到负链的数据 Reads map to ‘-’ |
|---|---|---|---|---|---|---|
| H0-1 | 42794280 | 35486057 | 31690935 | 3795122 | 16073999 | 16721352 |
| H0-2 | 43715068 | 36362495 | 32008861 | 4353634 | 15860703 | 16888759 |
| H0-3 | 39139946 | 32347565 | 28590712 | 3756853 | 14374026 | 15045589 |
| H8-1 | 46080164 | 37362124 | 33004646 | 4357478 | 16597665 | 17394638 |
| H8-2 | 38666666 | 30216711 | 26296339 | 3920372 | 12726728 | 13856534 |
| H8-3 | 52664656 | 43972928 | 39050460 | 4922468 | 19761602 | 20580516 |
| H24-1 | 36239984 | 28099032 | 24677641 | 3421391 | 12290831 | 13074940 |
| H24-2 | 78358258 | 63574337 | 55516961 | 8057376 | 27856971 | 29552246 |
| H24-3 | 41094422 | 34030303 | 30035515 | 3994788 | 15052963 | 15928683 |
| 差异表达基因集名称DEG set | 差异表达基因DEGs | 上调基因Up-regulated | 下调基因Down-regulated |
|---|---|---|---|
| H8 vs H0 | 5065 | 2442 | 2623 |
| H24 vs H8 | 2293 | 938 | 1355 |
| H24 vs H0 | 9022 | 4279 | 4743 |
表4 差异表达基因数目统计
Table 4 Statistic of number of differentially expressed genes
| 差异表达基因集名称DEG set | 差异表达基因DEGs | 上调基因Up-regulated | 下调基因Down-regulated |
|---|---|---|---|
| H8 vs H0 | 5065 | 2442 | 2623 |
| H24 vs H8 | 2293 | 938 | 1355 |
| H24 vs H0 | 9022 | 4279 | 4743 |
名称 DEG set | 总数 Total | 直系同源蛋白数据库COG | 基因本体数据库GO | 京都基因和基因组途径数据库百科全书KEGG | 真核直系同源基因数据库KOG | 非冗余蛋白质序列数据库Nr | 蛋白质家族数据库Pfam | 蛋白质序列数据库Swiss-Prot |
|---|---|---|---|---|---|---|---|---|
| H8 vs H0 | 5043 | 1787 | 4181 | 3550 | 2701 | 4956 | 4059 | 3970 |
| H24 vs H8 | 2279 | 1019 | 1920 | 1614 | 1248 | 2271 | 1855 | 1811 |
| H24 vs H0 | 8959 | 3247 | 7404 | 6317 | 4984 | 8861 | 7072 | 6900 |
表5 各数据库注释差异表达基因数量统计
Table 5 Statistic of the number of differentially expressed genes annotated in each database
名称 DEG set | 总数 Total | 直系同源蛋白数据库COG | 基因本体数据库GO | 京都基因和基因组途径数据库百科全书KEGG | 真核直系同源基因数据库KOG | 非冗余蛋白质序列数据库Nr | 蛋白质家族数据库Pfam | 蛋白质序列数据库Swiss-Prot |
|---|---|---|---|---|---|---|---|---|
| H8 vs H0 | 5043 | 1787 | 4181 | 3550 | 2701 | 4956 | 4059 | 3970 |
| H24 vs H8 | 2279 | 1019 | 1920 | 1614 | 1248 | 2271 | 1855 | 1811 |
| H24 vs H0 | 8959 | 3247 | 7404 | 6317 | 4984 | 8861 | 7072 | 6900 |
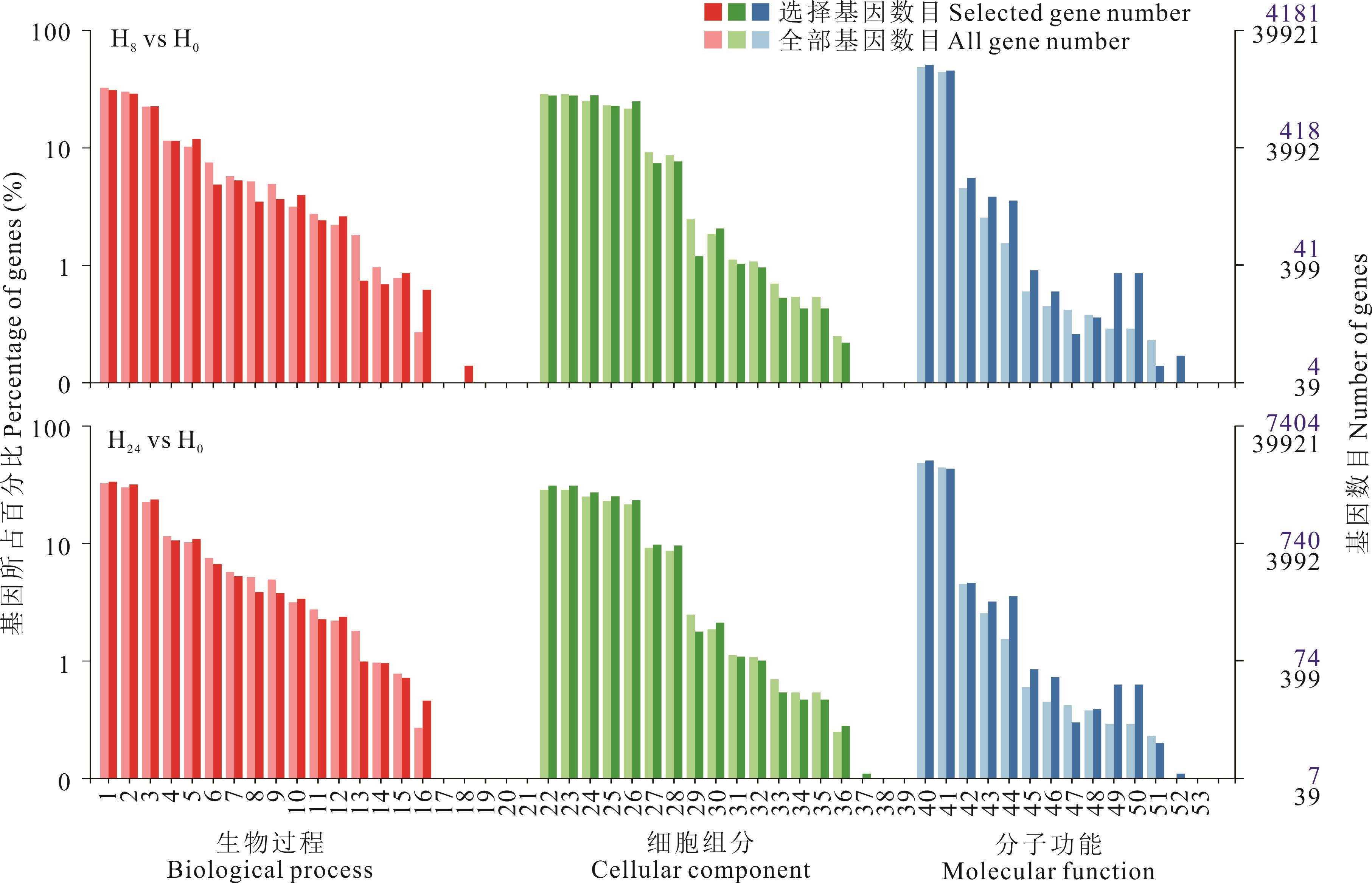
图2 差异表达基因GO注释分类统计1: 细胞过程Cellular process; 2: 代谢过程Metabolic process; 3: 单一生物过程Single-organism process; 4: 生物调节Biological regulation; 5: 刺激响应Response to stimulus; 6: 细胞组成或生物发生Cellular component or biogenesis; 7: 定位Localization; 8: 发育过程Developmental process; 9: 多细胞生物过程Multicellular organismal process; 10: 信号转导Signaling; 11: 生殖过程Reproductive process; 12: 多元生物过程Multi-organism process; 13: 繁殖Reproduction; 14: 生长Growth; 15: 免疫系统Immune system; 16: 节律过程Rhythmic process; 17: 运动Locomotion; 18: 生物黏附Biological adhesion; 19:解毒作用Detoxification; 20: 行为Behavior; 21: 细胞聚集Cell aggregation; 22: 细胞Cell; 23: 细胞组分Cell part; 24: 膜Membrane; 25: 细胞器Organelle; 26: 膜组分Membrane part; 27: 细胞器组分Organelle part; 28: 大分子复合物Macromolecular complex; 29: 膜封闭腔Membrane-enclosed lumen; 30: 胞外区域Extracellular region; 31: 细胞连接Cell junction; 32: 共质体Symplasm; 33: 胞外区域部分Extracellular region part; 34: 其他生物Other organism; 35: 其他生物组分Other organism part; 36: 超分子复合体Supramolecular complex; 37: 类核Nucleoid; 38: 突触Synapse; 39: 突触部分Synapse part; 40: 催化活性Catalytic activity; 41: 结合Binding; 42: 转运体活性Transporter activity; 43: 核酸结合转录因子活性Nucleic acid binding transcription factor activity; 44: 结构分子活性Structural molecular activity; 45: 抗氧化活性Antioxidant activity; 46: 电子载体活性Electron carrier activity; 47: 转录因子活性Transcription factor activity; 48:蛋白结合Protein binding; 49: 分子功能监管机构Molecular function regulator; 50: 分子换能器活性Molecular transducer activity; 51: 信号传感器活动Signal transducer activity; 52: 养分库活性Nutrient reservoir activity; 53: 蛋白质标记Protein tag.
Fig.2 Statistics of GO annotation classification of differentially expressed genes

图3 差异表达基因KEGG富集气泡点的大小代表该通路中注释的差异表达基因数,点的颜色代表超几何检验的P值。The size of the dots represents the number of annotated differentially expressed genes in the pathway, and the color of the dots represents the P-value of the hypergeometric test.
Fig.3 KEGG enrichment bubble of differentially expressed genes

图4 植物-病原互作通路相关基因在淹水胁迫不同时间点的表达模式
Fig.4 Expression patterns of genes related to plant-pathogen interaction pathways at different time points under waterlogging stress
代谢通路 Pathway | 基因ID Gene ID | 基因名称 Gene name | 差异倍数log2FC | 功能注释 Functional annotation | |
|---|---|---|---|---|---|
| H8 vs H0 | H24 vs H0 | ||||
植物-病原互作 Plant-pathogen interaction | |||||
Plant hormone signal transduction | |||||
碳代谢 Carbon metabolism | L195_g032111 | ||||
乙醛酸和二羧酸 代谢 Glyoxylate and dicarboxylate metabolism | |||||
表6 核心通路中差异表达基因功能注释统计
Table 6 Statistic of functional annotation of differentially expressed genes in core pathways
代谢通路 Pathway | 基因ID Gene ID | 基因名称 Gene name | 差异倍数log2FC | 功能注释 Functional annotation | |
|---|---|---|---|---|---|
| H8 vs H0 | H24 vs H0 | ||||
植物-病原互作 Plant-pathogen interaction | |||||
Plant hormone signal transduction | |||||
碳代谢 Carbon metabolism | L195_g032111 | ||||
乙醛酸和二羧酸 代谢 Glyoxylate and dicarboxylate metabolism | |||||

图5 植物激素信号通路相关基因在淹水胁迫不同时间点的表达模式
Fig.5 Expression patterns of plant hormone signal transduction pathway related genes at different time points under waterlogging stress

图7 乙醛酸和二羧酸代谢通路相关基因在淹水胁迫不同时间点的表达模式
Fig.7 Expression patterns of glyoxylate and dicarboxylate metabolism pathway related genes at different time points under waterlogging stress
转录因子 TF | 对比 Compare | 差异表达基因DEGs | 上调基因Up-regulated | 下调基因 Down-regulated | 主要通路富集 Primary pathway enrichment |
|---|---|---|---|---|---|
| AP2/ERF | H8 vs H0 | 4 | 4 | 0 | 糖酵解/糖异生、 戊糖磷酸途径、果糖和甘露糖代谢、碳代谢、氨基酸生物合成、RNA降解 Glycolysis/gluconeogenesis,pentose phosphate pathway,fructose and mannose metabolism,carbon metabolism,biosynthesis of amino acids,RNA degradation |
| H24 vs H0 | 6 | 5 | 1 | ||
| WRKY | H8 vs H0 | 30 | 6 | 24 | 剪接体、MAPK信号通路-植物、植物-病原互作Spliceosome,MAPK signaling pathway-plant,plant-pathogen interaction |
| H24 vs H0 | 44 | 16 | 28 | ||
| bHLH | H8VS H0 | 34 | 13 | 21 | 植物激素信号转导、昼夜节律-植物、MAPK信号通路-植物、植物-病原互作Plant hormone signal transduction,circadian rhythm-plant,MAPK signaling pathway-plant,plant-pathogen interaction |
| H24 vs H0 | 46 | 16 | 30 | ||
| NAC | H8 vs H0 | 62 | 22 | 40 | 核糖体、MAPK信号通路-植物、RNA转运、淀粉和蔗糖代谢、植物激素信号转导Ribosome,MAPK signaling pathway-plant,RNA transport,starch and sucrose metabolism,plant hormone signal transduction |
| H24 vs H0 | 145 | 41 | 104 | ||
| bZIP | H8 vs H0 | 13 | 8 | 5 | RNA转运、植物激素信号转导、内质网中的蛋白质加工、MAPK信号通路-植物RNA transport,plant hormone signal transduction,protein processing in endoplasmic reticulum,MAPK signaling pathway-plant |
| H24 vs H0 | 27 | 18 | 9 |
表7 核心转录因子差异表达基因数量统计
Table 7 Statistic of number of differentially expressed genes of core transcription factors (TF)
转录因子 TF | 对比 Compare | 差异表达基因DEGs | 上调基因Up-regulated | 下调基因 Down-regulated | 主要通路富集 Primary pathway enrichment |
|---|---|---|---|---|---|
| AP2/ERF | H8 vs H0 | 4 | 4 | 0 | 糖酵解/糖异生、 戊糖磷酸途径、果糖和甘露糖代谢、碳代谢、氨基酸生物合成、RNA降解 Glycolysis/gluconeogenesis,pentose phosphate pathway,fructose and mannose metabolism,carbon metabolism,biosynthesis of amino acids,RNA degradation |
| H24 vs H0 | 6 | 5 | 1 | ||
| WRKY | H8 vs H0 | 30 | 6 | 24 | 剪接体、MAPK信号通路-植物、植物-病原互作Spliceosome,MAPK signaling pathway-plant,plant-pathogen interaction |
| H24 vs H0 | 44 | 16 | 28 | ||
| bHLH | H8VS H0 | 34 | 13 | 21 | 植物激素信号转导、昼夜节律-植物、MAPK信号通路-植物、植物-病原互作Plant hormone signal transduction,circadian rhythm-plant,MAPK signaling pathway-plant,plant-pathogen interaction |
| H24 vs H0 | 46 | 16 | 30 | ||
| NAC | H8 vs H0 | 62 | 22 | 40 | 核糖体、MAPK信号通路-植物、RNA转运、淀粉和蔗糖代谢、植物激素信号转导Ribosome,MAPK signaling pathway-plant,RNA transport,starch and sucrose metabolism,plant hormone signal transduction |
| H24 vs H0 | 145 | 41 | 104 | ||
| bZIP | H8 vs H0 | 13 | 8 | 5 | RNA转运、植物激素信号转导、内质网中的蛋白质加工、MAPK信号通路-植物RNA transport,plant hormone signal transduction,protein processing in endoplasmic reticulum,MAPK signaling pathway-plant |
| H24 vs H0 | 27 | 18 | 9 |
| 1 | Chen M J, Jia S X. Chinese forage plants. Beijing: China Agriculture Press, 2002. |
| 陈默君, 贾慎修. 中国饲用植物. 北京: 中国农业出版社, 2002. | |
| 2 | Jia S X. Forage plants of China-Vol.6. Beijing: China Agriculture Press, 1997. |
| 贾慎修. 中国饲用植物志-第六卷. 北京: 中国农业出版社, 1997. | |
| 3 | Chen B S. Forage crop cultivation. Beijing: China Agriculture Press, 2001. |
| 陈宝书. 牧草饲料作物栽培学. 北京: 中国农业出版社, 2001. | |
| 4 | Meng X J, Yu L P, Cheng W D, et al. Influence of rhizobia inoculation and nitrogen fertilizer application on isoflavonoides content of Minshan red clover. Pratacultural Science, 2010, 27(5): 97-100. |
| 孟祥君, 俞联平, 程文定, 等. 接种根瘤菌与施肥对岷山红三叶异黄酮含量的影响. 草业科学, 2010, 27(5): 97-100. | |
| 5 | Yuan J B, Lu J Z. Determination of soybean isoflavone with ultra-violet spectrophotometry. Soybean Science, 2004(2): 147-150. |
| 袁金斌, 卢建中. 紫外分光光度法测定大豆总异黄酮的含量. 大豆科学, 2004(2): 147-150. | |
| 6 | Wang Z M, Yue M Q, Du W H, et al. Excellent legume for feeding and medicine-Minshan Trifolium pratense. Pratacultural Science, 2005, 22(4): 33-35. |
| 王志明, 岳民勤, 杜文华, 等. 集饲用和药用价值于一体的牧草新秀-岷山红三叶. 草业科学, 2005, 22(4): 33-35. | |
| 7 | Phelan P, Moloney A P, McGeough E J, et al. Forage legumes for grazing and conserving in ruminant production systems. Critical Reviews in Plant Sciences, 2015, 34(4): 281-326. |
| 8 | Mckenna P, Cannon N, Conway J, et al. Red clover (Trifolium pratense) in conservation agriculture: A compelling case for increased adoption. International Journal of Agricultural Sustainability, 2018, 16: 342-366. |
| 9 | Tan S D, Zhu M Y, Zhang K R, et al. Response and adaptation of plants to submergence stress. Chinese Journal of Ecology, 2009, 28(9): 1871-1877. |
| 谭淑端, 朱明勇, 张克荣, 等. 植物对水淹胁迫的响应与适应. 生态学杂志, 2009, 28(9): 1871-1877. | |
| 10 | Bodegom P M V, Sorrell B K, Oosthoek A, et al. Separating the effects of partial submergence and soil oxygen demand on plant physiology. Ecology, 2008, 89(1): 193-204. |
| 11 | Silvertown J, Dodd M E, Gowing D, et al. Hydrologically defined niches reveal a basis for species richness in plant communities. Nature, 1999, 400: 61-63. |
| 12 | Normile D. Reinventing rice to feed the world. Science, 2008, 321: 330-333. |
| 13 | Mcmanmon M, Crawford R M. Metabolic theory of flooding tolerance-significance of enzyme distribution and behaviour. New Phytologist, 1971, 70(2): 299-306. |
| 14 | Visser E, Voesenek L. Acclimation to soil flooding-sensing and signal-transduction. Plant and Soil, 2005, 274(1/2): 197-214. |
| 15 | Voesenek L A C J, Sasidharan R. Ethylene- and oxygen signalling- drive plant survival during flooding. Plant Biology, 2013, 15(3): 426-435. |
| 16 | Zhang F S. Environmental stress and plant rhizosphere nutrition. Beijing: China Agriculture Press, 1998. |
| 张福锁. 环境胁迫与植物根际营养. 北京: 中国农业出版社, 1998. | |
| 17 | Klok E J, Wilson I W, Wilson D, et al. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell, 2002, 14(10): 2481-2494. |
| 18 | Zhang J, Tang L, Zhang Y J, et al. Application of transcriptome sequencing technique in the study of waterlogging stress in plant. Molecular Plant Breeding, 2019, 17(4): 1191-1202. |
| 张健, 唐露, 张雅洁, 等. 转录组测序技术在植物淹水胁迫研究中的应用. 分子植物育种, 2019, 17(4): 1191-1202. | |
| 19 | Loucks C E S, Deen W, Gaudin A C M, et al. Genotypic differences in red clover (Trifolium pratense) response under severe water deficit. Plant and Soil, 2018, 425(1/2): 401-414. |
| 20 | He W, Fan Y, Wang L, et al. Analysis of the grey incidence of wild Trifolium pratense drought resistance in the three gorges reservoir area. Acta Prataculturae Sinica, 2009, 18(3): 255-259. |
| 何玮, 范彦, 王琳, 等. 三峡库区野生红三叶苗期抗旱性灰色关联分析. 草业学报, 2009, 18(3): 255-259. | |
| 21 | Pu X J, Tian J S, Tian X H, et al. Construction of the AFLP linkage map and QTL analysis of powdery mildew resistance in red clover. Acta Prataculturae Sinica, 2018, 27(4): 79-88. |
| 蒲小剑, 田久胜, 田新会, 等. 红三叶遗传图谱构建及抗白粉病基因QTL定位. 草业学报, 2018, 27(4): 79-88. | |
| 22 | Zhang H S, Gao Q, Zhang T T, et al. Comprehensive evaluation of copper tolerance of 30 germplasm resources of red clover (Trifolium pratense). Acta Prataculturae Sinica, 2021, 30(12): 117-128. |
| 张鹤山, 高秋, 张婷婷, 等. 30份红三叶种质资源耐铜性综合评价. 草业学报, 2021, 30(12): 117-128. | |
| 23 | Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics, 2009, 10(1): 57-63. |
| 24 | Ren S, Sun M, Yan H, et al. Identification and distribution of NBS-encoding resistance genes of Dactylis glomerata L. and its expression under abiotic and biotic stress. Biochemical Genetics, 2020, 58(6): 824-847. |
| 25 | Qiao D D, Zhang Y J, Xiong X M, et al. Transcriptome analysis on responses of orchardgrass (Dactylis glomerata L.) leaves to a short term flooding. Hereditas, 2020, 157(1): 1-16. |
| 26 | Zeng B, Zhang Y, Zhang A, et al. Transcriptome profiling of two Dactylis glomerata L. cultivars with different tolerance in response to submergence stress. Phytochemistry, 2020, 175: 112378. |
| 27 | Reis-Filho J S. Next-generation sequencing. Breast Cancer Research, 2009, 11(3): 1-7. |
| 28 | Pertea M, Pertea G M, Antonescu C M, et al. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 2015, 33(3): 290-295. |
| 29 | Love M I, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 2014, 15(12): 550. |
| 30 | Altschul S F, Madden T L, Schaffer A A, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 1997, 25(17): 3389-3402. |
| 31 | Ashburner M, Ball C A, Blake J A, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 2000, 25(1): 25-29. |
| 32 | Minoru K, Susumu G, Shuichi K, et al. The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004, 32: 277-280. |
| 33 | Uniprot C T. UniProt: The universal protein knowledgebase. Nucleic Acids Research, 2018, 32: 115-119. |
| 34 | Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021. Nucleic Acids Research, 2021, 49: 412-419. |
| 35 | Muller J, Szklarczyk D, Julien P, et al. egg NOG v2.0: Extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Research, 2010, 38: 190-195. |
| 36 | Bettler E, Krause R, Horn F, et al. NRSAS: Nuclear receptor structure analysis servers. Nucleic Acids Research, 2003, 31(13): 3400-3403. |
| 37 | Pruitt K D, Tatusova T, Maglott D R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research, 2007, 35: 61-65. |
| 38 | Tatusov R L, Galperin M Y, Natale D A, et al. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1): 33-36. |
| 39 | Ichimura K, Shinozaki K, Tena G, et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends in Plant Science, 2002, 7(7): 301-308. |
| 40 | Zhang H. Functional analysis of C2H2-type zinc finger protein ZFP182 and ZFP36 in ABA-induced antioxidant defense in rice. Nanjing: Nanjing Agricultural University, 2012. |
| 张宏. 水稻C2H2型锌指蛋白ZFP182和ZFP36在ABA诱导的抗氧化防护中的功能分析. 南京: 南京农业大学, 2012. | |
| 41 | Dahl C C, Baldwin I T. Deciphering the role of ethylene in plant-herbivore interactions. Journal of Plant Growth Regulation, 2007, 26(2): 201-209. |
| 42 | Loon L C, Geraats B P J, Linthorst H J M. Ethylene as a modulator of disease resistance in plants. Trends in Plant Science, 2006, 11(4): 184-191. |
| 43 | Pozo M J, Van Loon L C, Pieterse C. Jasmonates-signals in plant-microbe interactions. Trends in Plant Science, 2004, 23(3): 211-222. |
| 44 | Aslam S, Gul N, Mir M A, et al. Role of jasmonates, calcium, and glutathione in plants to combat abiotic stresses through precise signaling cascade. Frontiers in Plant Science, 2021, 12: 668029. |
| 45 | Hu X J, Mao D H. Genome-wide expression analysis of the phytohormones signalling pathway in rice seedlings by using RNA-Seq. Research of Agricultural Modernization, 2019, 40(5): 878-890. |
| 胡潇婕, 毛东海. 基于RNA-Seq技术分析植物激素信号途径在水稻幼苗中对低温胁迫的应答规律. 农业现代化研究, 2019, 40(5): 878-890. | |
| 46 | Gao H X, Zhu L, Liu T Q, et al. Transcriptomic analysis of plant hormone response to low temperature stress in rice. Molecular Plant Breeding, 2021, 19(13): 4188-4197. |
| 高红秀, 朱琳, 刘天奇, 等. 水稻植物激素响应低温胁迫反应的转录组分析. 分子植物育种, 2021, 19(13): 4188-4197. | |
| 47 | Zhou W, Yang Y, Zheng C, et al. Flooding represses soybean seed germination by mediating anaerobic respiration, glycometabolism and phytohormones biosynthesis. Environmental and Experimental Botany, 2021, 188: 104491. |
| 48 | Li Y, Shi L, Yang J, et al. Physiological and transcriptional changes provide insights into the effect of root waterlogging on the aboveground part of Pterocarya stenoptera. Genomics, 2021, 113(4): 2583-2590. |
| 49 | Wang X, He Y, Zhang C, et al. Physiological and transcriptional responses of Phalaris arundinacea under waterlogging conditions. Journal of Plant Physiology, 2021, 261(9): 153428. |
| 50 | Park S, Kim Y, Lee C, et al. Comparative transcriptome profiling of two sweet potato cultivars with contrasting flooding stress tolerance levels. Plant Biotechnology Reports, 2020, 14(6): 743-756. |
| 51 | Bromke M A. Amino acid biosynthesis pathways in diatoms. Metabolites, 2013, 3(2): 294-311. |
| 52 | Yalage D S M, Gambetta J M, Steel C C, et al. Elucidating the interaction of carbon, nitrogen, and temperature on the biosynthesis of Aureobasidium pullulans antifungal volatiles. Environmental Microbiology Reports, 2021, 13(4): 482-494. |
| 53 | Wang L, Yang Y R, Liu Y Q, et al. Transcriptome analysis of Chysanthemum×grandiflora in salt stress based on high-through-put sequencing. Molecular Plant Breeding, 2020, 18(5): 1419-1427. |
| 王琳, 杨伊如, 刘艳秋, 等. 基于高通量测序的露地菊(Chysanthemum×grandiflora)盐胁迫转录组分析. 分子植物育种, 2020, 18(5): 1419-1427. | |
| 54 | Qi X. The analysis of the differentially expressed genes in alfalfaunder cold stress at transcriptome level. Beijing: Chinese Academy of Agricultural Sciences, 2017. |
| 齐晓. 紫花苜蓿在转录组水平响应低温胁迫的差异表达基因研究. 北京: 中国农业科学院, 2017. | |
| 55 | Kramer R. Genetic and physiological approaches for the production of amino acids. Journal of Biotechnology, 1996, 45(1): 1-21. |
| 56 | Li Y H. Study on the regulation of metabolic pathway in aromatic amino acids biosynthesis. Beijing: Academy of Military Medical Sciences, 2003. |
| 李永辉. 芳香族氨基酸生物合成代谢途径调控研究. 北京: 中国人民解放军军事医学科学院, 2003. | |
| 57 | Lu Y. Analyses of rice genome sequences and expressed sequence tags and studies of glyoxylate cycle in submerged seedlings in rice. Shanghai: University of Chinese Academy of Sciences (Shanghai Institutes for Biological Sciences), 2006. |
| 陆颖. 水稻基因组和EST序列的测定和分析以及水淹条件下乙醛酸循环相关基因功能的研究. 上海: 中国科学院研究生院(上海生命科学研究院), 2006. | |
| 58 | Willekens H, Chamnongpol S, Davey M, et al. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. The EMBO Journal, 1997, 16(16): 4806-4816. |
| 59 | Mei Y, Chen L M. Research progresses on gene regulation and physiological role of plant formate dehygrogenase. China Biotechnology, 2010,30(5): 133-139. |
| 梅岩, 陈丽梅. 植物甲酸脱氢酶基因的表达调控及其生理功能研究进展. 中国生物工程杂志, 2010, 30(5): 133-139. |
| [1] | 杨志民, 邢瑞, 丁鋆嘉, 庄黎丽. 基于转录组测序的高羊茅分蘖与株高相关差异表达基因分析[J]. 草业学报, 2022, 31(1): 145-163. |
| [2] | 王诗雅, 郑殿峰, 冯乃杰, 梁喜龙, 项洪涛, 冯胜杰, 王新欣, 左官强. 鼓粒期淹水胁迫对大豆叶片AsA-GSH循环的损伤及烯效唑的缓解效应[J]. 草业学报, 2021, 30(7): 157-166. |
| [3] | 周晶, 陈思齐, 史文娇, 阳伏林, 林辉, 林占熺. 巨菌草幼叶及根转录组功能基因测序及分析[J]. 草业学报, 2021, 30(2): 143-155. |
| [4] | 张鹤山, 高秋, 张婷婷, 陆姣云, 田宏, 熊军波, 刘洋. 30份红三叶种质资源耐铜性综合评价[J]. 草业学报, 2021, 30(12): 117-128. |
| [5] | 汪芳珍, 杨成行, 何子华, 林子茹, 曾浩源, 马清. 盐处理下旱生植物沙芥蛋白激酶相关基因的差异表达分析[J]. 草业学报, 2021, 30(10): 116-124. |
| [6] | 钱晨, 刘智微, 钟小仙, 吴娟子, 张建丽, 潘玉梅. 海滨雀稗自交结实突变体及野生型幼穗组织的转录组分析[J]. 草业学报, 2019, 28(5): 132-142. |
| [7] | 成启明, 格根图, 撒多文, 王志军, 范文强, 卜振鲲, 司强, 李俊峰, 卢娟, 贾玉山. 不同品种紫花苜蓿转录组分析及营养品质差异的探讨[J]. 草业学报, 2019, 28(10): 199-208. |
| [8] | 李海云, 姚拓, 张榕, 张洁, 李智燕, 荣良燕, 路晓雯, 杨晓蕾, 夏东慧, 罗慧琴. 红三叶根际溶磷菌的筛选与培养基优化[J]. 草业学报, 2019, 28(1): 170-179. |
| [9] | 蒲小剑,田久胜,田新会,杜文华. 红三叶遗传图谱构建及抗白粉病基因QTL定位[J]. 草业学报, 2018, 27(4): 79-88. |
| [10] | 孟丽娟,赵桂琴. 国外引进红三叶种质在甘肃中部地区的生长特性及生产性能初步评价[J]. 草业学报, 2015, 24(9): 30-42. |
| [11] | 全瑞兰, 玉永雄. 淹水对紫花苜蓿南北方品种抗氧化酶和无氧呼吸酶的影响[J]. 草业学报, 2015, 24(5): 84-90. |
| [12] | 荣良燕,姚拓,马文彬,李德明,李儒仁,张洁,陆飒. 岷山红三叶根际优良促生菌对其宿主生长和品质的影响[J]. 草业学报, 2014, 23(5): 231-240. |
| [13] | 姜义宝,杨玉荣,冯长松,王成章,崔国文. 红三叶异黄酮对低温诱发肉鸡腹水综合症发生和抗氧化性能的影响[J]. 草业学报, 2013, 22(2): 94-99. |
| [14] | 俞联平,郝正里,李发弟,孟祥君,陈兴荣,李新媛. 岷山红三叶异黄酮对去卵巢大鼠生长和免疫及抗氧化指标的影响[J]. 草业学报, 2012, 21(6): 137-144. |
| [15] | 王瑞,梁坤伦,周志宇,郭霞,刘雪云 . 不同淹水梯度对紫穗槐的营养生长和生理响应[J]. 草业学报, 2012, 21(1): 149-155. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||