

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (1): 181-194.DOI: 10.11686/cyxb2021237
• 研究论文 • 上一篇
吴彤( ), 刘云苗, 金军, 董伟峰, 才晓溪, 孙明哲, 贾博为(
), 刘云苗, 金军, 董伟峰, 才晓溪, 孙明哲, 贾博为( ), 孙晓丽(
), 孙晓丽( )
)
收稿日期:2021-06-17
修回日期:2021-07-19
出版日期:2021-12-01
发布日期:2021-12-01
通讯作者:
贾博为,孙晓丽
作者简介:Corresponding author. E-mail: csmbl2016@126.com基金资助:
Tong WU( ), Yun-miao LIU, Jun JIN, Wei-feng DONG, Xiao-xi CAI, Ming-zhe SUN, Bo-wei JIA(
), Yun-miao LIU, Jun JIN, Wei-feng DONG, Xiao-xi CAI, Ming-zhe SUN, Bo-wei JIA( ), Xiao-li SUN(
), Xiao-li SUN( )
)
Received:2021-06-17
Revised:2021-07-19
Online:2021-12-01
Published:2021-12-01
Contact:
Bo-wei JIA,Xiao-li SUN
摘要:
CPA(cation proton antiporter)超家族通过转运质子和一价金属离子调节细胞内离子和pH稳定。阳离子质子转运体(cation/H+ exchanger,CHX)基因家族属于CPA2(cation proton antiporter 2)超家族,其N端有一个Na+/H+ exchanger结构域,对植物维持细胞离子平衡、花器官发育起着至关重要的作用。通过生物信息学手段,系统地分析了蒺藜苜蓿CHX家族基因,通过全基因组筛选共鉴定出47个MtCHXs;染色体定位分析表明蒺藜苜蓿CHX基因分布在8条不同的染色体上;同源性分析显示蒺藜苜蓿和拟南芥亲缘关系近,而与水稻亲缘关系远;进一步进化关系分析将47个MtCHXs基因分为5组,并且各组内成员在基因结构和基序上比较保守;顺式作用元件分析发现MtCHXs基因启动子包含大量光响应元件、激素响应元件以及干旱、低温、创伤响应元件;表达特性分析发现MtCHXs在生殖器官中高表达,并且响应干旱、低温等非生物胁迫。
吴彤, 刘云苗, 金军, 董伟峰, 才晓溪, 孙明哲, 贾博为, 孙晓丽. 蒺藜苜蓿cation/H+ exchanger基因家族鉴定及表达特征分析[J]. 草业学报, 2022, 31(1): 181-194.
Tong WU, Yun-miao LIU, Jun JIN, Wei-feng DONG, Xiao-xi CAI, Ming-zhe SUN, Bo-wei JIA, Xiao-li SUN. Identification and expression characteristics of a cation/H+ exchanger gene family in Medicago truncatula[J]. Acta Prataculturae Sinica, 2022, 31(1): 181-194.
序号 Number | 基因名 Gene name | 基因ID Gene ID | 序列长度Sequence length | 跨膜结构数目 No. of transmembrane | 预测蛋白定位 Predicted location | |||
|---|---|---|---|---|---|---|---|---|
基因序列 DNA (bp) | 转录本 Transcript (bp) | 编码区 CDS (bp) | 蛋白 Protein (aa) | |||||
| 1 | MtCHX1 | Medtr3g110700 | 2524 | 2358 | 2358 | 785 | 11 | PM |
| 2 | MtCHX2 | Medtr4g029520 | 2904 | 2367 | 2367 | 788 | 12 | PM |
| 3 | MtCHX3.1 | Medtr5g088450 | 3246 | 2415 | 2415 | 804 | 9 | PM |
| 4 | MtCHX3.2 | Medtr5g088470 | 3050 | 2397 | 2397 | 798 | 12 | PM |
| 5 | MtCHX3.3 | Medtr5g088430 | 3255 | 2406 | 2406 | 801 | 13 | PM |
| 6 | MtCHX3.4 | Medtr3g083850 | 3152 | 2394 | 2394 | 797 | 12 | PM |
| 7 | MtCHX3.5 | Medtr6g015940 | 2913 | 2373 | 2373 | 790 | 8 | PM |
| 8 | MtCHX3.6 | Medtr8g085260 | 2880 | 2385 | 2385 | 794 | 12 | PM |
| 9 | MtCHX3.7 | Medtr3g498715 | 2851 | 2397 | 2397 | 798 | 9 | PM |
| 10 | MtCHX3.8 | Medtr5g088790 | 3175 | 2466 | 2466 | 821 | 10 | PM |
| 11 | MtCHX3.9 | Medtr7g052895 | 2980 | 2190 | 2190 | 729 | 10 | PM |
| 12 | MtCHX3.10 | Medtr7g053000 | 3148 | 2466 | 2466 | 821 | 9 | PM |
| 13 | MtCHX4.1 | Medtr5g088840 | 2805 | 2466 | 2466 | 821 | 9 | PM |
| 14 | MtCHX4.2 | Medtr7g052680 | 3177 | 2466 | 2466 | 821 | 9 | PM |
| 15 | MtCHX4.3 | Medtr7g052970 | 3148 | 2466 | 2466 | 821 | 10 | PM |
| 16 | MtCHX6a.1 | Medtr6g053320 | 4103 | 2840 | 2319 | 772 | 10 | PM |
| 17 | MtCHX13 | Medtr3g031160 | 3075 | 2367 | 2367 | 788 | 10 | PM |
| 18 | MtCHX14.1 | Medtr5g074360 | 3383 | 2463 | 2463 | 820 | 11 | PM |
| 19 | MtCHX14.2 | Medtr5g074380 | 3932 | 2580 | 2580 | 859 | 11 | PM |
| 20 | MtCHX14.3 | Medtr5g074340 | 4814 | 2472 | 2472 | 823 | 9 | PM |
| 21 | MtCHX15.1 | Medtr3g071990 | 3746 | 2933 | 2442 | 813 | 12 | PM |
| 22 | MtCHX15.2 | Medtr3g031170 | 3612 | 2400 | 2400 | 799 | 10 | PM |
| 23 | MtCHX15.3 | Medtr5g074320 | 2612 | 2256 | 2256 | 751 | 10 | PM |
| 24 | MtCHX15.4 | Medtr3g071870 | 3200 | 2484 | 2484 | 827 | 10 | PM |
| 25 | MtCHX15.5 | Medtr3g071880 | 3169 | 2466 | 2466 | 821 | 10 | PM |
| 26 | MtCHX15.6 | Medtr2g011520 | 3139 | 2442 | 2442 | 813 | 12 | PM |
| 27 | MtCHX15.7 | Medtr3g096380 | 2801 | 2619 | 2517 | 838 | 12 | PM |
| 28 | MtCHX15.8 | Medtr8g093780 | 2741 | 2496 | 2496 | 831 | 12 | PM |
| 29 | MtCHX18.1 | Medtr5g009780 | 3692 | 2683 | 2412 | 803 | 10 | PM |
| 30 | MtCHX18.2 | Medtr5g009800 | 3026 | 2322 | 2322 | 773 | 9 | PM |
| 31 | MtCHX18.3 | Medtr5g009770 | 5043 | 4085 | 2403 | 800 | 12 | PM |
| 32 | MtCHX19.1 | Medtr4g078120 | 3509 | 2460 | 2460 | 819 | 10 | PM |
| 33 | MtCHX19.2 | Medtr4g123583 | 7284 | 2591 | 2436 | 811 | 10 | PM |
| 34 | MtCHX20.1 | Medtr7g099820 | 6199 | 2683 | 2439 | 812 | 9 | PM |
| 35 | MtCHX20.2 | Medtr7g099750 | 8579 | 3300 | 2535 | 844 | 11 | PM |
| 36 | MtCHX20.3 | Medtr7g099800 | 6317 | 3111 | 2556 | 851 | 10 | PM |
| 37 | MtCHX23.1 | Medtr3g037270 | 4511 | 2448 | 2448 | 815 | 10 | PM |
| 38 | MtCHX23.2 | Medtr3g037260 | 2973 | 2442 | 2442 | 813 | 11 | PM |
| 39 | MtCHX23.3 | Medtr5g067690 | 3624 | 2451 | 2451 | 816 | 10 | PM |
| 40 | MtCHX23.4 | Medtr0002s1180 | 3901 | 2433 | 2433 | 810 | 11 | PM |
| 41 | MtCHX23.5 | Medtr1g045870 | 3637 | 2454 | 2379 | 792 | 10 | PM |
| 42 | MtCHX23.6 | Medtr1g043400 | 3728 | 2601 | 2424 | 807 | 10 | PM |
| 43 | MtCHX23.7 | Medtr2g040100 | 3664 | 2523 | 2523 | 840 | 8 | PM |
| 44 | MtCHX23.8 | Medtr7g085870 | 3367 | 2496 | 2496 | 831 | 10 | PM |
| 45 | MtCHX25 | Medtr3g465680 | 3108 | 2763 | 2763 | 920 | 10 | PM |
| 46 | MtCHX28.1 | Medtr4g037755 | 2447 | 2358 | 2358 | 785 | 10 | PM |
| 47 | MtCHX28.2 | Medtr3g031030 | 3350 | 2361 | 2361 | 786 | 11 | PM |
表 1 蒺藜苜蓿CHX基因家族成员信息
Table 1 Information of CHX family genes from M. truncatula
序号 Number | 基因名 Gene name | 基因ID Gene ID | 序列长度Sequence length | 跨膜结构数目 No. of transmembrane | 预测蛋白定位 Predicted location | |||
|---|---|---|---|---|---|---|---|---|
基因序列 DNA (bp) | 转录本 Transcript (bp) | 编码区 CDS (bp) | 蛋白 Protein (aa) | |||||
| 1 | MtCHX1 | Medtr3g110700 | 2524 | 2358 | 2358 | 785 | 11 | PM |
| 2 | MtCHX2 | Medtr4g029520 | 2904 | 2367 | 2367 | 788 | 12 | PM |
| 3 | MtCHX3.1 | Medtr5g088450 | 3246 | 2415 | 2415 | 804 | 9 | PM |
| 4 | MtCHX3.2 | Medtr5g088470 | 3050 | 2397 | 2397 | 798 | 12 | PM |
| 5 | MtCHX3.3 | Medtr5g088430 | 3255 | 2406 | 2406 | 801 | 13 | PM |
| 6 | MtCHX3.4 | Medtr3g083850 | 3152 | 2394 | 2394 | 797 | 12 | PM |
| 7 | MtCHX3.5 | Medtr6g015940 | 2913 | 2373 | 2373 | 790 | 8 | PM |
| 8 | MtCHX3.6 | Medtr8g085260 | 2880 | 2385 | 2385 | 794 | 12 | PM |
| 9 | MtCHX3.7 | Medtr3g498715 | 2851 | 2397 | 2397 | 798 | 9 | PM |
| 10 | MtCHX3.8 | Medtr5g088790 | 3175 | 2466 | 2466 | 821 | 10 | PM |
| 11 | MtCHX3.9 | Medtr7g052895 | 2980 | 2190 | 2190 | 729 | 10 | PM |
| 12 | MtCHX3.10 | Medtr7g053000 | 3148 | 2466 | 2466 | 821 | 9 | PM |
| 13 | MtCHX4.1 | Medtr5g088840 | 2805 | 2466 | 2466 | 821 | 9 | PM |
| 14 | MtCHX4.2 | Medtr7g052680 | 3177 | 2466 | 2466 | 821 | 9 | PM |
| 15 | MtCHX4.3 | Medtr7g052970 | 3148 | 2466 | 2466 | 821 | 10 | PM |
| 16 | MtCHX6a.1 | Medtr6g053320 | 4103 | 2840 | 2319 | 772 | 10 | PM |
| 17 | MtCHX13 | Medtr3g031160 | 3075 | 2367 | 2367 | 788 | 10 | PM |
| 18 | MtCHX14.1 | Medtr5g074360 | 3383 | 2463 | 2463 | 820 | 11 | PM |
| 19 | MtCHX14.2 | Medtr5g074380 | 3932 | 2580 | 2580 | 859 | 11 | PM |
| 20 | MtCHX14.3 | Medtr5g074340 | 4814 | 2472 | 2472 | 823 | 9 | PM |
| 21 | MtCHX15.1 | Medtr3g071990 | 3746 | 2933 | 2442 | 813 | 12 | PM |
| 22 | MtCHX15.2 | Medtr3g031170 | 3612 | 2400 | 2400 | 799 | 10 | PM |
| 23 | MtCHX15.3 | Medtr5g074320 | 2612 | 2256 | 2256 | 751 | 10 | PM |
| 24 | MtCHX15.4 | Medtr3g071870 | 3200 | 2484 | 2484 | 827 | 10 | PM |
| 25 | MtCHX15.5 | Medtr3g071880 | 3169 | 2466 | 2466 | 821 | 10 | PM |
| 26 | MtCHX15.6 | Medtr2g011520 | 3139 | 2442 | 2442 | 813 | 12 | PM |
| 27 | MtCHX15.7 | Medtr3g096380 | 2801 | 2619 | 2517 | 838 | 12 | PM |
| 28 | MtCHX15.8 | Medtr8g093780 | 2741 | 2496 | 2496 | 831 | 12 | PM |
| 29 | MtCHX18.1 | Medtr5g009780 | 3692 | 2683 | 2412 | 803 | 10 | PM |
| 30 | MtCHX18.2 | Medtr5g009800 | 3026 | 2322 | 2322 | 773 | 9 | PM |
| 31 | MtCHX18.3 | Medtr5g009770 | 5043 | 4085 | 2403 | 800 | 12 | PM |
| 32 | MtCHX19.1 | Medtr4g078120 | 3509 | 2460 | 2460 | 819 | 10 | PM |
| 33 | MtCHX19.2 | Medtr4g123583 | 7284 | 2591 | 2436 | 811 | 10 | PM |
| 34 | MtCHX20.1 | Medtr7g099820 | 6199 | 2683 | 2439 | 812 | 9 | PM |
| 35 | MtCHX20.2 | Medtr7g099750 | 8579 | 3300 | 2535 | 844 | 11 | PM |
| 36 | MtCHX20.3 | Medtr7g099800 | 6317 | 3111 | 2556 | 851 | 10 | PM |
| 37 | MtCHX23.1 | Medtr3g037270 | 4511 | 2448 | 2448 | 815 | 10 | PM |
| 38 | MtCHX23.2 | Medtr3g037260 | 2973 | 2442 | 2442 | 813 | 11 | PM |
| 39 | MtCHX23.3 | Medtr5g067690 | 3624 | 2451 | 2451 | 816 | 10 | PM |
| 40 | MtCHX23.4 | Medtr0002s1180 | 3901 | 2433 | 2433 | 810 | 11 | PM |
| 41 | MtCHX23.5 | Medtr1g045870 | 3637 | 2454 | 2379 | 792 | 10 | PM |
| 42 | MtCHX23.6 | Medtr1g043400 | 3728 | 2601 | 2424 | 807 | 10 | PM |
| 43 | MtCHX23.7 | Medtr2g040100 | 3664 | 2523 | 2523 | 840 | 8 | PM |
| 44 | MtCHX23.8 | Medtr7g085870 | 3367 | 2496 | 2496 | 831 | 10 | PM |
| 45 | MtCHX25 | Medtr3g465680 | 3108 | 2763 | 2763 | 920 | 10 | PM |
| 46 | MtCHX28.1 | Medtr4g037755 | 2447 | 2358 | 2358 | 785 | 10 | PM |
| 47 | MtCHX28.2 | Medtr3g031030 | 3350 | 2361 | 2361 | 786 | 11 | PM |

图6 蒺藜苜蓿CHX基因家族保守性分析A: 蒺藜苜蓿CHX基因家族保守结构域The conserved domain of the CHX gene family in alfalfa; B: 蒺藜苜蓿CHX基因家族保守基序The conserved motif of the CHX gene family in alfalfa.
Fig.6 Conservation analysis of MtCHXs
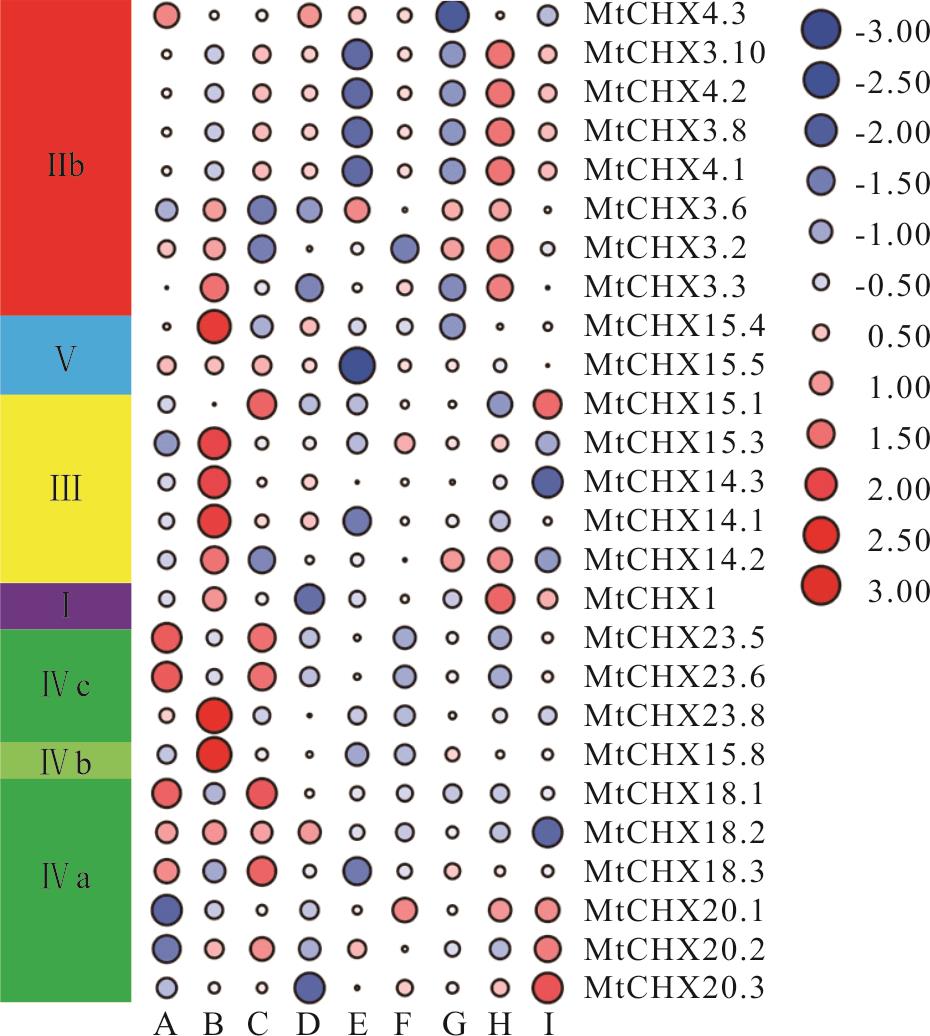
图9 蒺藜苜蓿CHX基因家族组织表达特性分析A: 种子 Seed; B: 花 Flower; C: 根瘤 Nodule; D: 荚 Pod; E: 营养芽 Vegetative bud; F: 叶柄 Petiole; G: 茎 Stem; H: 叶 Leaf; I: 根 Root.
Fig.9 Expression profile of MtCHXs genes in different tissues
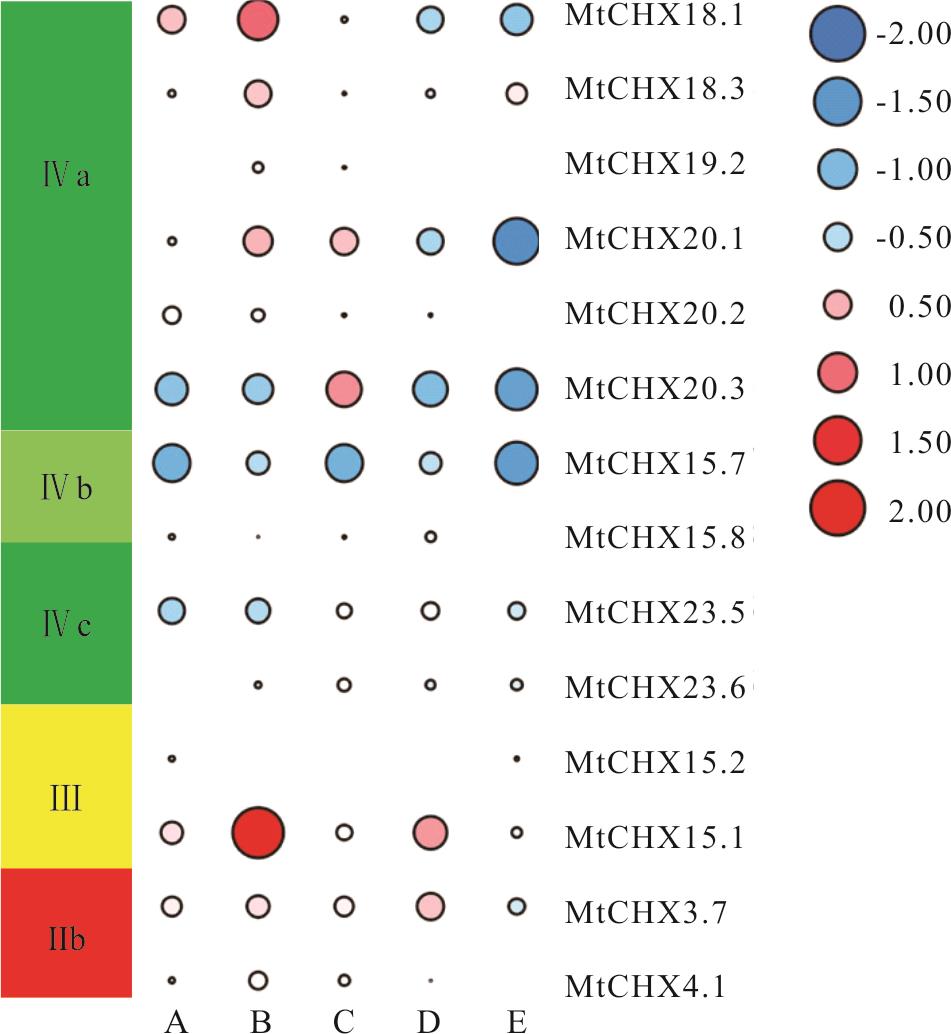
图10 蒺藜苜蓿CHX基因家族非生物胁迫表达模式分析A: 冷害 Cold; B: 冻害 Freezing; C: 干旱胁迫 Drought; D: 盐胁迫 Salt; E: 脱落酸处理 ABA.
Fig.10 Expression pattern of MtCHXs genes in response to various abiotic stress
| 1 | Wang Z H. Cultivation techniques of alfalfa. Technical Advisor for Animal Husbandry, 2021(4): 53-55. |
| 王兆辉. 苜蓿的种植技术. 现代畜牧科技, 2021(4): 53-55. | |
| 2 | Geng H, Li X Q, Li H X, et al. Study on ecological adaptability of introduced alfalfa varieties in midwestern Jilin Province. Journal of Northeast Agricultural Sciences, 2021(7): 1-7. |
| 耿慧, 李晓秋, 李鸿祥, 等. 吉林省中西部地区引进苜蓿品种生态适应性研究. 东北农业科学, 2021(7): 1-7. | |
| 3 | Guo L Z, Hu H J. Advances in studies on potassium circulation and recirculation in plants. Journal of Gansu Agricultural University, 2001(1): 1-7. |
| 郭丽琢, 胡恒觉. 植物体内钾循环与再循环的研究进展. 甘肃农业大学学报, 2001(1): 1-7. | |
| 4 | Shen Y, Cai X X, Jia B W, et al. Studies on the interaction between wild soybean cationic proton transporter CHX19.3 and 14-3-3 protein. Journal of Heilongjiang Bayi Agricultural University, 2020, 32(1): 1-9. |
| 沈阳, 才晓溪, 贾博为, 等. 野生大豆阳离子质子转运体CHX19.3与14-3-3蛋白的相互作用研究. 黑龙江八一农垦大学学报, 2020, 32(1): 1-9. | |
| 5 | Chanroj S, Wang G Y, Venema K, et al. Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Frontiers in Plant Science, 2012, 3: 25. |
| 6 | Sze H, Padmanaban S, Cellier F, et al. Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiology, 2004, 136(1): 2532-2547. |
| 7 | Zhao J, Li P H, Motes C M, et al. CHX14 is a plasma membrane K-efflux transporter that regulates K+ redistribution in Arabidopsis thaliana. Plant, Cell & Environment, 2015, 38(11): 2223-2238. |
| 8 | Zhao J, Cheng N H, Motes C M, et al. AtCHX13 is a plasma membrane K+ transporter. Plant Physiology, 2008, 148(2): 796-807. |
| 9 | Chanroj S, Lu Y X, Padmanaban S, et al. Plant-specific cation/H+ exchanger 17 and its homologs are endomembrane K+ transporters with roles in protein sorting. Journal of Biological Chemistry, 2011, 286(39): 33931-33941. |
| 10 | Padmanaban S, Chanroj S, Kwak J M, et al. Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiology, 2007, 144(1): 82-93. |
| 11 | Cai X X, Shen Y, Zhou W H, et al. Genome-wide identification and bioinformatics analysis of soybean CHX gene family. Genomics and Applied Biology, 2018, 37(12): 5360-5369. |
| 才晓溪, 沈阳, 周伍红, 等. 大豆CHX基因家族全基因组鉴定与生物信息学分析. 基因组学与应用生物学, 2018, 37(12): 5360-5369. | |
| 12 | Jia B W, Sun M Z, Duanmu H Z, et al. GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Scientific Reports, 2017, 7(1): 9423. |
| 13 | Chen Y, Ma J K, Miller A J, et al. OsCHX14 is involved in the K+ homeostasis in rice (Oryza sativa) flowers. Plant & Cell Physiology, 2016, 57(7): 1-14. |
| 14 | Yang L, Dong L, Li M J, et al. Identification and expression analysis of K+ transporter gene family in apple. Acta Horticulturae Sinica, 2016, 43(6): 1021-1032. |
| 杨琳, 董玲, 李明军, 等. 苹果K+转运蛋白基因家族鉴定及表达分析. 园艺学报, 2016, 43(6): 1021-1032. | |
| 15 | Goodstein D M, Shu S Q, Howson R, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Research, 2012, 40(D1): D1178-D1186. |
| 16 | Sara E G, Jaina M, Alex B A, et al. The Pfam protein families database in 2019. Nucleic Acids Research, 2019, 47(D1): D427-D432. |
| 17 | Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research, 2018, 46(D1): D493-D496. |
| 18 | Marchler B A, Bryant S H. CD-Search: protein domain annotations on the fly. Nucleic Acids Research, 2004, 32(Issue suppl_2): 327-331. |
| 19 | Kenta N, Paul H. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in Biochemical Sciences, 1999, 24(1): 34-36. |
| 20 | Chen C, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 21 | Xu L, Dong Z B, Fang L, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research, 2019, 47(W1): 52-58. |
| 22 | Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. |
| 23 | Bailey T L, Boden M, Buske F A, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Research, 2009, 37(Issue suppl_2): 202-208. |
| 24 | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 2002, 30(1): 325-327. |
| 25 | Waese J, Fan J, Pasha A, et al. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. The Plant Cell, 2017, 29(8): 1806-1821. |
| 26 | Wang L G. Advances in studies on Na+, K+/H+ reverse transporters in Arabidopsis thaliana. Chinese Journal of Biotechnology, 2019, 35(8): 1424-1432. |
| 王立光. 拟南芥内膜Na+, K+/H+反向转运体研究进展. 生物工程学报, 2019, 35(8): 1424-1432. | |
| 27 | Zheng S, Pan T, Fan L G, et al. A novel AtKEA gene family, homolog of bacterial K+/H+ antiporters, plays potential roles in K+ homeostasis and osmotic adjustment in Arabidopsis. PLoS One, 2013, 8(11): e81463. |
| 28 | Mottaleb S A, Rodríguez-Navarro A, Haro R. Knockouts of Physcomitrella patens CHX1 and CHX2 transporters reveal high complexity of potassium homeostasis. Plant and Cell Physiology, 2013, 54(9): 1455-1468. |
| 29 | Shi Q L. The significance of developing green ecological agriculture in our country. Forest By-Product and Speciality in China, 2013(2): 101-102. |
| 石庆龙. 我国大力发展绿色生态化广义农业的意义. 中国林副特产, 2013(2): 101-102. | |
| 30 | Xiang J H, Song T T, Yuan Y Y, et al. Bioinformatics and expression analysis of MsGT6 in alfalfa under stress. Molecular Plant Breeding, 2020, 18(23): 7742-7750. |
| 项继红, 宋婷婷, 袁玉莹, 等. 紫花苜蓿MsGT6的生物信息学及其逆境胁迫下表达分析. 分子植物育种, 2020, 18(23): 7742-7750. | |
| 31 | Blondon F, Marie D, Brown S, et al. Genome size and base composition in Medicago sativa and M. truncatula species. NRC Research Press, 1994, 37(2): 264-270. |
| 32 | Liu W W, Cui H T, Wei C X, et al. Cloning and functional analysis of chlorophyllide a oxygenase encoding gene MtCAO in Medicago truncatula. Acta Prataculturae Sinica, 2020, 29(5): 171-184. |
| 刘文文, 崔会婷, 尉春雪, 等. 蒺藜苜蓿叶绿素酸酯a加氧酶(MtCAO)基因的克隆与功能分析. 草业学报, 2020, 29(5): 174-184. | |
| 33 | Huang S Y, Hu T M, Yang P Z, et al. Identification and functional analysis of the PYL gene family in Medicago truncatula. Pratacultural Science, 2019, 36(2): 422-423. |
| 黄思源, 呼天明, 杨培志, 等. 蒺藜苜蓿PYL基因家族的全基因组鉴定、表达和功能分析. 草业科学, 2019, 36(2): 422-423. | |
| 34 | Li F, He X H, Zhang Y B, et al. Genome-wide identification and analysis of the TCP transcription factor family of Medicago truncatula. Molecular Plant Breeding, 2018, 16(20): 6639-6645. |
| 李菲, 何小红, 张宇斌, 等. 蒺藜苜蓿TCP转录因子家族的全基因组鉴定和分析. 分子植物育种, 2018, 16(20): 6639-6645. | |
| 35 | Yang Q, Niu X C, Wang R G, et al. Genome-wide characterization of DUF221 gene family in Medicago truncatula and screening for salt response genes. Mocecular Plant Breeding, 2019, 17(16): 5255-5262 |
| 杨杞, 牛肖翠, 王瑞刚, 等. 蒺藜苜蓿DUF221基因家族全基因组鉴定及盐响应相关基因筛选. 分子植物育种, 2019, 17(16): 5255-5262. | |
| 36 | Song Z H, Niu L L, Yang Q, et al. Genome-wide identification and characterization of UGT family in pigeonpea (Cajanus cajan) and expression analysis in abiotic stress. Trees, 2019, 33(4): 987-1002. |
| 37 | Sonnhammer E, Koonin E V. Orthology, paralogy and proposed classification for paralog subtypes. Trends in Genetics, 2002, 18(12): 619-620. |
| 38 | Qiao Y G, Chen L, Wang Y F, et al. Screening and expression analysis of Dof gene family in medicinal plant Lonicerae japonica. Journal of Nuclear Agricultural Sciences, 2020, 34(9): 1889-1897. |
| 乔永刚, 陈亮, 王勇飞, 等. 药用植物金银花Dof基因家族的筛选及表达分析. 核农学报, 2020, 34(9): 1889-1897. | |
| 39 | Yin H, Yuan Y Y, Song T T, et al. Bioinformatics analysis of the YABBY gene family in alfalfa (Medicago sativa) and its response to stress. Molecular Plant Breeding, 2020, 18(2): 416-424. |
| 尹航, 袁玉莹, 宋婷婷, 等. 紫花苜蓿(Medicago sativa)YABBY基因家族的生物信息学分析及其对逆境胁迫的响应. 分子植物育种, 2020, 18(2): 416-424. | |
| 40 | Song J, Cao X N, Wang H G, et al. Identification and expression analysis of SBP protein gene family in foxtail millet. Journal of Nuclear Agricultural Sciences, 2020, 34(7): 1409-1420. |
| 宋健, 曹晓宁, 王海岗, 等. 谷子SBP蛋白基因家族的鉴定及表达分析. 核农学报, 2020, 34(7): 1409-1420. | |
| 41 | Ma C, Song P, Shang S S, et al. Identification and expression pattern analysis of GRFs gene family in Brachypodium distachyon. Journal of Nuclear Agricultural Sciences, 2020, 34(6): 1152-1162. |
| 马超, 宋鹏, 尚申申, 等. 二穗短柄草GRFs基因家族的鉴定及表达模式分析. 核农学报, 2020, 34(6): 1152-1162. | |
| 42 | Liu J J, She L L, Lan X Z, et al. Bioinformatics and expression analysis of MYB gene family based on transcriptome of cold-acclimated Mirabilis himalaica callus. Chinese Agricultural Science Bulletin, 2020, 36(29): 54-61. |
| 刘佳佳, 佘露露, 兰小中, 等. 喜马拉雅紫茉莉MYB家族的生物信息学及表达分析. 中国农学通报, 2020, 36(29): 54-61. | |
| 43 | Chen J, Fang Y P, Jiang J M, et al. KNOX gene family identification and expression analysis of SbKNOX22 gene in Sorghum. Journal of Nuclear Agricultural Sciences, 2020, 34(11): 2377-2385. |
| 陈俊, 方远鹏, 蒋君梅, 等. 高粱KNOX基因家族鉴定及SbKNOX22基因表达分析. 核农学报, 2020, 34(11): 2377-2385. | |
| 44 | Liu Y J, Feng Y M, Liu X Y, et al. Effect of cadmium on seed germination of Persian Chrysanthemum and migrant effect of exogenous MeJA. Acta Prataculturae Sinica, 2017, 26(1): 122-130. |
| 刘宇婧, 冯艺玫, 刘欣悦, 等. 镉对波斯菊种子发芽的影响及外源MeJA的缓解作用. 草业学报, 2017, 26(1): 122-130. | |
| 45 | Zhang Y X, Cong B M, Wang X G, et al. Correlation analysis of cold resistance of alfalfa and antioxidant enzyme activity in roots. Acta Agrestia Sinica, 2021, 29(2): 244-249. |
| 张玉霞, 丛百明, 王显国, 等. 苜蓿抗寒性与根系抗氧化酶活性相关性分析. 草地学报, 2021, 29(2): 244-249. | |
| 46 | Chu Y, Ren F, Zhao G L, et al. Research progress on the effect of osmotic stress on antioxidant enzymes in plants. Journal of Anhui Agricultural Sciences, 2011, 39(3): 1300-1302. |
| 褚妍, 任菲, 赵贵林, 等. 渗透胁迫对植物抗氧化酶影响的研究进展. 安徽农业科学, 2011, 39(3): 1300-1302. | |
| 47 | Du Y Y, Lian H J, Li X M, et al. Genome-wide identification and bioinformatics analysis of grape CHX gene family. Molecular Plant Breeding, 2020, 18(24): 8009-8021. |
| 杜宜洋, 连红娟, 李晓梅, 等. 葡萄CHX基因家族全基因组鉴定及生物信息学分析. 分子植物育种, 2020, 18(24): 8009-8021. |
| [1] | 刘文文, 崔会婷, 尉春雪, 龙瑞才, 康俊梅, 杨青川, 王珍. 蒺藜苜蓿叶绿素酸酯a加氧酶(MtCAO)基因的克隆与功能分析[J]. 草业学报, 2020, 29(5): 171-181. |
| [2] | 张智琦, 王珍, 张铁军, 龙瑞才, 杨青川, 康俊梅. 蒺藜苜蓿MtNSN1的克隆与功能分析[J]. 草业学报, 2019, 28(8): 200-208. |
| [3] | 王小山, 季晓敏, 刘隆阳, 纪冰沁, 田银芳. EBR对NaCl胁迫下苜蓿属植物离子吸收和分配的影响[J]. 草业学报, 2018, 27(9): 110-119. |
| [4] | 史经昂, 张兵, 肖晓琳, 马晶晶, 杨向阳, 刘建秀. 结缕草肉桂醇脱氢酶基因家族全基因组序列鉴定和表达分析[J]. 草业学报, 2017, 26(6): 111-119. |
| [5] | 任海龙, 魏臻武, 陈祥. 蒺藜苜蓿、天蓝苜蓿、金花菜基因组SNP穿梭标记开发[J]. 草业学报, 2017, 26(4): 188-195. |
| [6] | 张雪, 孙鑫博, 樊波, 张胤冰, 韩烈保, 许立新. 结缕草ZjCSD基因的克隆及表达分析[J]. 草业学报, 2017, 26(2): 102-110. |
| [7] | 邵麟惠, 郑兴卫, 李聪. 蒺藜苜蓿E3泛素连接酶U-box基因克隆及表达分[J]. 草业学报, 2016, 25(7): 62-72. |
| [8] | 郑兴卫, 邵麟惠, 李聪. 蒺藜苜蓿全基因组中U-box基因家族的筛选与特征分析[J]. 草业学报, 2015, 24(8): 130-141. |
| [9] | 刘文瑜, 杨宏伟, 魏小红, 刘博, 王高强, 吴伟涛. 外源NO调控盐胁迫下蒺藜苜蓿种子萌发生理特性及抗氧化酶的研究[J]. 草业学报, 2015, 24(2): 85-95. |
| [10] | 杨国锋,苏昆龙,赵怡然,宋智斌,孙娟. 蒺藜苜蓿叶绿体密码子偏好性分析[J]. 草业学报, 2015, 24(12): 171-179. |
| [11] | 张军,宋丽莉,郭东林,郭长虹,束永俊. MADS-box基因家族在蒺藜苜蓿的全基因组分析[J]. 草业学报, 2014, 23(6): 233-241. |
| [12] | 邹雪,张烨,吴明阳,王西瑶. 马铃薯和拟南芥GAPC酶基因的克隆及分析[J]. 草业学报, 2014, 23(1): 239-247. |
| [13] | 王丽,张俊莲,张金文,刘玉汇,白江平,余斌,杨宏羽,王蒂. 拟南芥高亲和性K+载体蛋白基因cDNA克隆及其序列特征分析[J]. 草业学报, 2013, 22(6): 230-238. |
| [14] | 姜格格,宋丽莉,郭东林,蔡洪生,郭长虹,束永俊. 蒺藜苜蓿耐酸铝性状的全基因组关联分析[J]. 草业学报, 2013, 22(4): 170-178. |
| [15] | 李剑,张金林,王锁民,郭强. 小花碱茅HKT2;1基因全长cDNA的克隆与生物信息学分析[J]. 草业学报, 2013, 22(2): 140-149. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||