

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (6): 150-162.DOI: 10.11686/cyxb2021130
• 研究论文 • 上一篇
杨兴云1( ), 乔丹丹1, 张雅洁1, 王少青2, 任俊才1, 李明阳1, 屈明好1, 尚盼盼1, 杨成1, 黄琳凯3,4(
), 乔丹丹1, 张雅洁1, 王少青2, 任俊才1, 李明阳1, 屈明好1, 尚盼盼1, 杨成1, 黄琳凯3,4( ), 曾兵1,2(
), 曾兵1,2( )
)
收稿日期:2021-04-07
修回日期:2021-07-07
出版日期:2022-06-20
发布日期:2022-05-11
通讯作者:
黄琳凯,曾兵
作者简介:E-mail: huanglinkai@siau.edu.cn基金资助:
Xing-yun YANG1( ), Dan-dan QIAO1, Ya-jie ZHANG1, Shao-qing WANG2, Jun-cai REN1, Ming-yang LI1, Ming-hao QU1, Pan-pan SHANG1, Cheng YANG1, Lin-kai HUANG3,4(
), Dan-dan QIAO1, Ya-jie ZHANG1, Shao-qing WANG2, Jun-cai REN1, Ming-yang LI1, Ming-hao QU1, Pan-pan SHANG1, Cheng YANG1, Lin-kai HUANG3,4( ), Bing ZENG1,2(
), Bing ZENG1,2( )
)
Received:2021-04-07
Revised:2021-07-07
Online:2022-06-20
Published:2022-05-11
Contact:
Lin-kai HUANG,Bing ZENG
摘要:
水淹胁迫是限制我国西南地区鸭茅产量和品质提升的主要环境因子,已经成为一种不容忽视的非生物胁迫。鉴定鸭茅耐涝相关的功能基因,并探究其调控机制是鸭茅种质创新,提高鸭茅耐涝能力的必要途径。以鸭茅耐涝品种“滇北”为试验材料,分别经水淹胁迫处理0、8和24 h后,利用Illumina Hiseq测序平台对鸭茅叶片进行小RNA测序。结果表明,在水淹胁迫处理下共鉴定得到208个差异表达基因(DEGs),经过筛选后有38个基因上调表达,34个基因下调表达,共占差异表达基因的34.62%。“滇北”鸭茅在水淹胁迫下差异表达基因主要属于miR166、miR167、miR159、miR396和miR156这5个miRNA基因家族。基于对差异miRNA进行靶基因预测及靶基因的GO和KEGG功能分析,发现这些靶基因主要参与细胞生理过程、代谢过程、IL-17信号通路、Th17细胞分化等植物逆境响应过程,为进一步揭示鸭茅在水淹胁迫下的分子调控机制提供了研究线索。
杨兴云, 乔丹丹, 张雅洁, 王少青, 任俊才, 李明阳, 屈明好, 尚盼盼, 杨成, 黄琳凯, 曾兵. 鸭茅响应水淹胁迫的miRNA差异表达分析[J]. 草业学报, 2022, 31(6): 150-162.
Xing-yun YANG, Dan-dan QIAO, Ya-jie ZHANG, Shao-qing WANG, Jun-cai REN, Ming-yang LI, Ming-hao QU, Pan-pan SHANG, Cheng YANG, Lin-kai HUANG, Bing ZENG. A differential gene expression analysis of miRNA in Dactylis glomerata in response to flooding stress[J]. Acta Prataculturae Sinica, 2022, 31(6): 150-162.
样本 Sample | 质控前reads数 Total reads | 质控后reads数 Clean reads | 定位到基因组上的sRNA总数 Total mapped sRNA | Q20 (%) | Q30 (%) | GC (% ) |
|---|---|---|---|---|---|---|
| DB-0 h-1 | 13115501 | 6911279 | 690667 | 99.6 | 98.8 | 52.9 |
| DB-0 h-2 | 12506705 | 7412332 | 847591 | 99.8 | 99.3 | 52.1 |
| DB-0 h-3 | 13129146 | 6898821 | 711644 | 99.8 | 99.4 | 51.8 |
| DB-8 h-1 | 10196159 | 5869117 | 1032354 | 99.5 | 98.7 | 51.5 |
| DB-8 h-2 | 12254067 | 7088163 | 1113861 | 99.2 | 98.1 | 51.2 |
| DB-8 h-3 | 11877997 | 7548708 | 1138457 | 99.6 | 99.0 | 51.6 |
| DB-24 h-1 | 11852644 | 8729342 | 1401312 | 99.8 | 99.3 | 52.0 |
| DB-24 h-2 | 12412282 | 8960959 | 1337594 | 99.8 | 99.6 | 51.6 |
| DB-24 h-3 | 12481155 | 9370332 | 1435461 | 99.8 | 99.3 | 52.1 |
表1 不同鸭茅样品测序数据统计
Table 1 Statistical results of sequencing data of different D. glomerata samples
样本 Sample | 质控前reads数 Total reads | 质控后reads数 Clean reads | 定位到基因组上的sRNA总数 Total mapped sRNA | Q20 (%) | Q30 (%) | GC (% ) |
|---|---|---|---|---|---|---|
| DB-0 h-1 | 13115501 | 6911279 | 690667 | 99.6 | 98.8 | 52.9 |
| DB-0 h-2 | 12506705 | 7412332 | 847591 | 99.8 | 99.3 | 52.1 |
| DB-0 h-3 | 13129146 | 6898821 | 711644 | 99.8 | 99.4 | 51.8 |
| DB-8 h-1 | 10196159 | 5869117 | 1032354 | 99.5 | 98.7 | 51.5 |
| DB-8 h-2 | 12254067 | 7088163 | 1113861 | 99.2 | 98.1 | 51.2 |
| DB-8 h-3 | 11877997 | 7548708 | 1138457 | 99.6 | 99.0 | 51.6 |
| DB-24 h-1 | 11852644 | 8729342 | 1401312 | 99.8 | 99.3 | 52.0 |
| DB-24 h-2 | 12412282 | 8960959 | 1337594 | 99.8 | 99.6 | 51.6 |
| DB-24 h-3 | 12481155 | 9370332 | 1435461 | 99.8 | 99.3 | 52.1 |
| 样本Sample | 数据库miRBase | 外显子Exon | 内含子Intron | 基因间Intergenic | 重复序列Repeat |
|---|---|---|---|---|---|
| DB-0 h | 11.00 | 30.81 | 13.00 | 95.46 | 99.6 |
| DB-8 h | 15.80 | 17.02 | 11.86 | 93.30 | 99.8 |
| DB-24 h | 14.00 | 22.14 | 8.73 | 90.51 | 99.5 |
表2 小RNA分类统计
Table 2 Small RNA classification statistics (%)
| 样本Sample | 数据库miRBase | 外显子Exon | 内含子Intron | 基因间Intergenic | 重复序列Repeat |
|---|---|---|---|---|---|
| DB-0 h | 11.00 | 30.81 | 13.00 | 95.46 | 99.6 |
| DB-8 h | 15.80 | 17.02 | 11.86 | 93.30 | 99.8 |
| DB-24 h | 14.00 | 22.14 | 8.73 | 90.51 | 99.5 |
| 类型Type | 数量Number | 上调Up | 下调Down |
|---|---|---|---|
| DB 0 h vs 8 h | 70 | 34 | 36 |
| DB 0 h vs 24 h | 66 | 32 | 34 |
| DB 8 h vs 24 h | 72 | 38 | 34 |
表3 差异基因统计
Table 3 Statistical of differentially expressed genes (DEGs)
| 类型Type | 数量Number | 上调Up | 下调Down |
|---|---|---|---|
| DB 0 h vs 8 h | 70 | 34 | 36 |
| DB 0 h vs 24 h | 66 | 32 | 34 |
| DB 8 h vs 24 h | 72 | 38 | 34 |
| DB 0 h vs 8 h | DB 0 h vs 24 h | DB 8 h vs 24 h |
|---|---|---|
| miR166 (28) | miR166 (20) | miR166 (30) |
| miR167 (9) | miR159 (13) | miR159 (13) |
| miR159 (9) | miR167 (9) | miR167 (11) |
| miR156 (8) | miR396 (7) | miR156 (7) |
| miR396 (5) | miR156 (6) | miR396 (5) |
表4 鸭茅中响应水淹胁迫的差异表达miRNA家族
Table 4 Differentially expressed miRNA families in D. glomerata in response to flooding stress
| DB 0 h vs 8 h | DB 0 h vs 24 h | DB 8 h vs 24 h |
|---|---|---|
| miR166 (28) | miR166 (20) | miR166 (30) |
| miR167 (9) | miR159 (13) | miR159 (13) |
| miR159 (9) | miR167 (9) | miR167 (11) |
| miR156 (8) | miR396 (7) | miR156 (7) |
| miR396 (5) | miR156 (6) | miR396 (5) |
序号 Number | miRNA名称 miRNA name | miRNA对应的靶基因 Target gene |
|---|---|---|
| 1 | nta-miR166c | Bradi3g28970.1; Bradi3g28970.2; Bradi1g47666.1; Bradi5g18830.1; Bradi3g51590.1; Bradi2g06210.1; Bradi3g15175.2 |
| 2 | gma-miR167f | Bradi4g02473.1; Bradi1g16570.2; Bradi4g01730.1; Bradi1g67570.1; Bradi1g16570.1 |
| 3 | ahy-miR160-5p | Bradi3g28950.1; Bradi5g15904.1; Bradi5g27400.1; Bradi5g27400.2; Bradi1g33160.1; Bradi3g49320.1 |
| 4 | mes-miR159a | Bradi1g36542.1; Bradi1g15581.1; Bradi2g06832.5; Bradi2g06832.4;Bradi2g06832.3;Bradi2g06832.2;Bradi2g06832.1; Bradi1g15581.3; Bradi5g17600.2; Bradi2g20670.1; Bradi1g15581.2; Bradi2g53010.1 |
| 5 | bdi-miR396a-5p | Bradi5g18961.1; Bradi1g12650.2; Bradi1g12650.1; Bradi3g52547.1; Bradi1g50597.1; Bradi4g16450.3; Bradi3g57267.1; Bradi1g09900.2; Bradi1g09900.1; Bradi5g20607.1; Bradi4g16450.2; Bradi1g05540.1; Bradi1g46427.2; Bradi1g464 27.1; Bradi3g51685.1 |
| 6 | ptc-miR393c | Bradi5g08680.1; Bradi2g35720.1 |
| 7 | sbi-miR166i | Bradi3g15175.2; Bradi3g28970.2; Bradi5g18830.1; Bradi3g51590.1; Bradi4g01887.1; Bradi3g28970.1; Bradi2g06210.1; Bradi1g47666.1; Bradi1g01640.2; Bradi1g13910.1 |
表5 不同的靶基因对应同一个miRNA
Table 5 Different target genes correspond to the same miRNA
序号 Number | miRNA名称 miRNA name | miRNA对应的靶基因 Target gene |
|---|---|---|
| 1 | nta-miR166c | Bradi3g28970.1; Bradi3g28970.2; Bradi1g47666.1; Bradi5g18830.1; Bradi3g51590.1; Bradi2g06210.1; Bradi3g15175.2 |
| 2 | gma-miR167f | Bradi4g02473.1; Bradi1g16570.2; Bradi4g01730.1; Bradi1g67570.1; Bradi1g16570.1 |
| 3 | ahy-miR160-5p | Bradi3g28950.1; Bradi5g15904.1; Bradi5g27400.1; Bradi5g27400.2; Bradi1g33160.1; Bradi3g49320.1 |
| 4 | mes-miR159a | Bradi1g36542.1; Bradi1g15581.1; Bradi2g06832.5; Bradi2g06832.4;Bradi2g06832.3;Bradi2g06832.2;Bradi2g06832.1; Bradi1g15581.3; Bradi5g17600.2; Bradi2g20670.1; Bradi1g15581.2; Bradi2g53010.1 |
| 5 | bdi-miR396a-5p | Bradi5g18961.1; Bradi1g12650.2; Bradi1g12650.1; Bradi3g52547.1; Bradi1g50597.1; Bradi4g16450.3; Bradi3g57267.1; Bradi1g09900.2; Bradi1g09900.1; Bradi5g20607.1; Bradi4g16450.2; Bradi1g05540.1; Bradi1g46427.2; Bradi1g464 27.1; Bradi3g51685.1 |
| 6 | ptc-miR393c | Bradi5g08680.1; Bradi2g35720.1 |
| 7 | sbi-miR166i | Bradi3g15175.2; Bradi3g28970.2; Bradi5g18830.1; Bradi3g51590.1; Bradi4g01887.1; Bradi3g28970.1; Bradi2g06210.1; Bradi1g47666.1; Bradi1g01640.2; Bradi1g13910.1 |
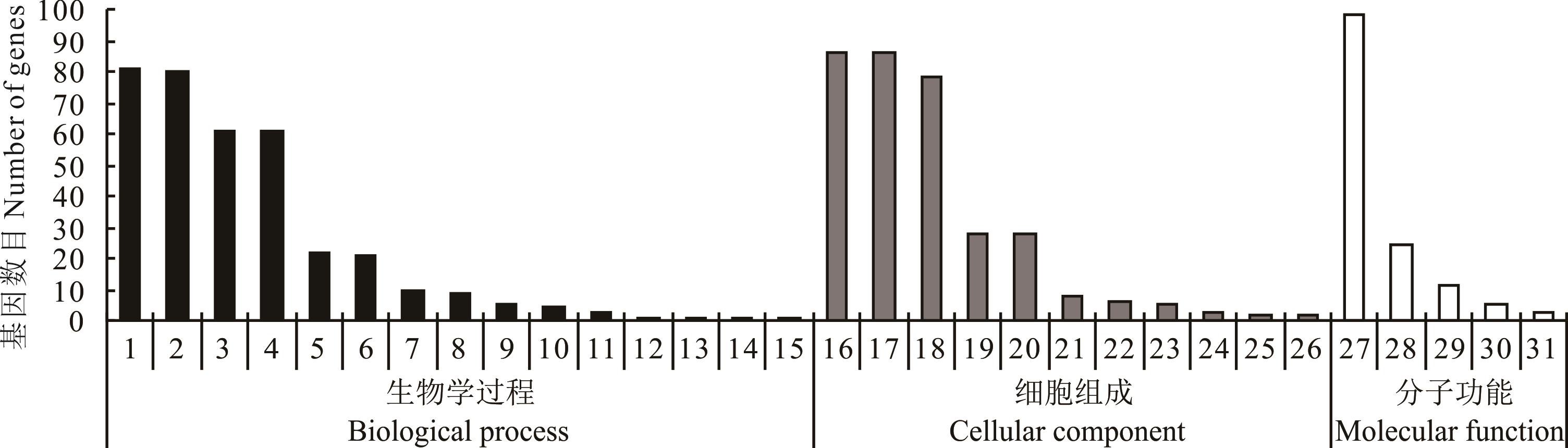
图1 “滇北”淹水0 h vs 8 h差异miRNA 靶基因GO分类1:细胞过程 Cellular process;2:代谢过程 Metabolic process;3:生物调控 Metabolic process;4:生物过程调控 Regulation of biological process;5:单一有机体过程 Single-organism process;6:发育过程 Developmental process;7:对刺激的反应 Response to stimulus;8:定位 Localization;9:生物过程的正向调节 Positive regulation of biological process;10:信号 Signaling;11:多细胞生物的过程 Multicellular organismal process;12:细胞组成组织或生物发生 Cellular component organization or biogenesis;13:生物过程的负调控 Negative regulation of biological process;14:繁殖 Reproduction;15:生殖过程 Reproductive process;16:细胞 Cell;17:细胞部分 Cell part;18:细胞器 Organelle;19:膜 Membrane;20:膜部分 Membrane part;21:细胞器部分 Organelle part;22:大分子复合体 Macromolecular complex;23:细胞外区域 Extracellular region;24:超分子络合物 Supramolecular complex;25:细胞连接Cell junction;26:合胞体 Symplast;27:黏合物 Binding;28:催化活性 Catalytic activity;29:核酸结合转录因子活性 Nucleic acid binding transcription factor activity;30:结构分子活性 Structural molecule activity;31:转运活力 Transporter activity.
Fig.1 Histogram of GO classification of differential miRNA target genes for 0 h vs 8 h flooding in “Dianbei”

图2 “滇北”淹水0 h vs 24 h差异miRNA 靶基因GO分类1:细胞过程 Cellular process;2:代谢过程 Metabolic process;3:生物调节Biological regulation;4:生物过程调控 Regulation of biological process; 5:单一有机体过程 Single-organism process;6:发育过程 Developmental process;7:对刺激的反应 Response to stimulus;8:定位 Localization;9:信号 Signaling;10:生物过程的正向调节 Positive regulation of biological process;11:多细胞生物的过程 Multicellular organismal process;12:细胞组成组织或生物发生 Cellular component organization or biogenesis;13:生物过程的负调控 Negative regulation of biological process;14:繁殖 Reproduction;15:生殖过程 Reproductive process;16:细胞 Cell;17:细胞部分 Cell part;18:细胞器 Organelle;19:膜 Membrane;20:膜部分 Membrane part;21:细胞器部分 Organelle part;22:高分子复合物 Macromolecular complex;23:超分子络合物 Supramolecular complex;24:细胞外区域 Extracellular region;25:黏合物 Binding;26:催化活性 Catalytic activity;27:核酸结合转录因子活性 Nucleic acid binding transcription factor activity;28:结构分子活性 Structural molecule activity;29:转运活力 Transporter activity.
Fig.2 Histogram of GO classification of differential miRNA target genes for 0 h vs 24 h flooding in “Dianbei”
| 1 | Lindner R, Garcia A. Geographic distribution and genetic resources of Dactylis in Galicia (Northwest Spain). Genetic Resources and Crop Evolution, 1997, 44(6): 499-507. |
| 2 | Madesis P, Abraham E M, Kalivas A, et al. Genetic diversity and structure of natural Dactylis glomerata L. populations revealed by morphological and microsatellite-based (SSR/ISSR) markers. Genetics and Molecular Research, 2014, 13(2): 4226-4240. |
| 3 | Last L, Widmer F, Fjellstad W, et al. Genetic diversity of natural orchardgrass (Dactylis glomerata L.) populations in three regions in Europe. BMC Genetics, 2013, 14(1): 1-14. |
| 4 | Ji Y, Chen P, Chen J, et al. Combinations of small RNA, RNA, and degradome sequencing uncovers the expression pattern of microRNA-mRNA pairs adapting to drought stress in leaf and root of Dactylis glomerata L. International Journal of Molecular Sciences, 2018, 19(10): 3114. |
| 5 | Huang L K, Yan H D, Zhao X X, et al. Identifying differentially expressed genes under heat stress and developing molecular markers in orchardgrass (Dactylis glomerata L.) through transcriptome analysis. Molecular Ecology Resources, 2015, 15(6): 1497-1509. |
| 6 | Yan H, Zhang Y, Zeng B, et al. Genetic diversity and association of EST-SSR and SCoT markers with rust traits in orchardgrass (Dactylis glomerata L). Molecules, 2016, 21(1): 66. |
| 7 | Kumutha D, Sairam R K, Ezhilmathi K, et al. Effect of waterlogging on carbohydrate metabolism in pigeon pea (Cajanus cajan L.): Upregulation of sucrose synthase and alcohol dehydrogenase. Plant Science, 2008, 175(5): 706-716. |
| 8 | Wang L, Zhang Y, Yang X. Research progress on waterlogging tolerance of vegetable crops. Chinese Vegetables, 2017(11): 14-20. |
| 王露, 张宇, 杨旭. 蔬菜作物耐涝性研究进展. 中国蔬菜, 2017(11): 14-20. | |
| 9 | Ren B C, Zhu Y L, Li X, et al. Effects of waterlogging on photosynthetic characteristics of summer maize under field conditions. Acta Agronomica Sinica, 2015, 41(2): 329-338. |
| 任佰朝, 朱玉玲, 李霞, 等. 大田淹水对夏玉米光合特性的影响. 作物学报, 2015, 41(2): 329-338. | |
| 10 | Wu X, Tang Y, Li C, et al. Chlorophyll fluorescence and yield responses of winter wheat to waterlogging at different growth stages. Plant Production Science, 2015, 18(3): 284-294. |
| 11 | Xue H, Pu Y, Meng Y, et al. Antioxidant responses to waterlogging stress and subsequent recovery in two Kentucky bluegrass (Poa pratensis L.) cultivars. Acta Physiologiae Plantarum, 2015, 37(10): 197. |
| 12 | Zhang J, Tang L, Zhang Y J, et al. Application of transcriptome sequencing technique in the study of waterlogging stress in plants. Molecular Plant Breeding, 2019, 17(4): 1191-1202. |
| 张健, 唐露, 张雅洁, 等. 转录组测序技术在植物水淹胁迫研究中的应用. 分子植物育种, 2019, 17(4): 1191-1202. | |
| 13 | Lai E C. MicroRNAs: Runts of the genome assert themselves. Current Biology, 2003, 13(23): 925-936. |
| 14 | Bartel D P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 2004, 116(2): 281-297. |
| 15 | Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell, 2009, 136(4): 669-687. |
| 16 | Zhang B. MicroRNA: A new target for improving plant tolerance to abiotic stress. Journal of Experimental Botany, 2015, 66(7): 1749-1761. |
| 17 | Kumar V, Khare T, Shriram V, et al. Plant small RNAs: The essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Reports, 2018, 37(1): 61-75. |
| 18 | Liu Q, Yan S, Yang T, et al. Small RNAs in regulating temperature stress response in plants. Journal of Integrative Plant Biology, 2017, 59(11): 774-791. |
| 19 | Paul S, Datta S K, Datta K. miRNA regulation of nutrient homeostasis in plants. Frontiers in Plant Science, 2015, 6: 232. |
| 20 | De la Rosa C, Covarrubias A A, Reyes J L. A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant, Cell & Environment, 2019, 42(1): 133-144. |
| 21 | Campbell M T, Proctor C A, Dou Y, et al. Genetic and molecular characterization of submergence response identifies Subtol6 as a major submergence tolerance locus in maize. PLoS One, 2015, 10(3): e0120385. |
| 22 | Liu Z, Kumari S, Zhang L, et al. Characterization of miRNAs in response to short-term waterlogging in three inbred lines of Zea mays. PLoS One, 2012, 7(6): e39786. |
| 23 | Huo D, Sun L, Li X, et al. Differential expression of miRNAs in the respiratory tree of the sea cucumber Apostichopus japonicus under hypoxia stress. G3: Genes, Genomes, Genetics, 2017, 7(11): 3681-3692. |
| 24 | Licausi F, Weits D A, Pant B D, et al. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytologist, 2011, 190(2): 442-456. |
| 25 | Liu Z J. Characterize the roles of miRNAs in responding to short-term waterlogging in Zea mays root. Wuhan: Huazhong Agricultural University, 2012. |
| 刘智捷. 玉米苗期渍水胁迫诱导根系miRNA快速应答机理的研究. 武汉: 华中农业大学, 2012. | |
| 26 | Jin Q, Xu Y, Mattson N, et al. Identification of submergence-responsive microRNAs and their targets reveals complex miRNA-mediated regulatory networks in lotus (Nelumbo nucifera Gaertn.). Frontiers in Plant Science, 2017, 8: 6. |
| 27 | Gao Y Z, Zong J Q, Chen J B, et al. Responses of 15 different warm season turfgrass germplasms to long-term submergence and waterlogging stress. Pratacultural Science, 2015, 32(3): 354-362. |
| 高艳芝, 宗俊勤, 陈静波, 等. 15份不同暖季型草坪草生长量对长期水淹和水涝胁迫的响应. 草业科学, 2015, 32(3): 354-362. | |
| 28 | Abiko T, Kotula L, Shiono K, et al. Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea nicaraguensis contribute to its waterlogging tolerance as compared with maize (Zea mays ssp. mays). Plant, Cell & Environment, 2012, 35(9): 1618-1630. |
| 29 | Shi M Z, Zhou B S. Research advance in physiological damage of flood and water-logging resistance. Anhui Agricultural Sciences, 2006(2): 209-210. |
| 时明芝, 周保松. 植物涝害和耐涝机理研究进展. 安徽农业科学, 2006(2): 209-210. | |
| 30 | Westra S, Fowler H J, Evans J P, et al. Future changes to the intensity and frequency of short-duration extreme rainfall. Reviews of Geophysics, 2015, 52(3): 522-555. |
| 31 | Klaas M, Haiminen N, Grant J, et al. Transcriptome characterization and differentially expressed genes under flooding and drought stress in the biomass grasses Phalaris arundinacea and Dactylis glomerata. Annals of Botany, 2019, 124(4): 717-730. |
| 32 | Zeng B, Zhang Y, Zhang A, et al. Transcriptome profiling of two Dactylis glomerata L. cultivars with different tolerance in response to submergence stress. Phytochemistry, 2020, 175: 112378. |
| 33 | Qiao D, Zhang Y, Xiong X, et al. Transcriptome analysis on responses of orchardgrass (Dactylis glomerata L.) leaves to a short term flooding. Hereditas, 2020, 157(1): 20. |
| 34 | Kozomara A, Griffiths-Jones S. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 2014, 42: 68-73. |
| 35 | Friedländer M R, Mackowiak S D, Li N, et al. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research, 2012, 40(1): 37-52. |
| 36 | Wu H J, Ma Y K, Chen T, et al. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Research, 2012, 40: 22-28. |
| 37 | Bo X, Wang S. TargetFinder: A software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics, 2005, 21(8): 1401-1402. |
| 38 | Langmead B, Salzberg S L. Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012, 9(4): 357-359. |
| 39 | Jones-Rhoades M W, Bartel D P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell, 2004, 14(6): 787-799. |
| 40 | Ren Y, Chen L, Zhang Y, et al. Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Functional & Integrative Genomics, 2012, 12(2): 327-339. |
| 41 | Nawrocki E P, Burge S W, Bateman A, et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Research, 2015, 43(1): 130-137. |
| 42 | Ashburner M, Ball C A, Blake J A, et al. Gene ontology: Tool for the unification of biology. Nature Genetics, 2000, 25(1): 25-29. |
| 43 | Klopfenstein D V, Zhang L, Pedersen B S, et al. GOATOOLS: A python library for gene ontology analyses. Scientific Reports, 2018, 8(1): 1-17. |
| 44 | Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004, 32: 277-280. |
| 45 | Zeng M, He S H, Li W H, et al. Differentialexpression of miRNAand functionoftarget genesin heteromorphicleavesof Populus euphratica. Journal of Beijing Forestry University, 2020, 42(6): 1-13. |
| 曾明, 何书航, 李文海, 等. 胡杨异形叶差异表达miRNA及其靶基因功能分析. 北京林业大学学报, 2020, 42(6): 1-13. | |
| 46 | Wu L L. Cloning and functional research of miR166 and its target gene in potato. Lanzhou: Gansu Agricultural University, 2016. |
| 武亮亮. 马铃薯miR166及其靶基因的克隆和功能研究. 兰州: 甘肃农业大学, 2016. | |
| 47 | Ohashi-Ito K, Fukuda H.HD-zip Ⅲ homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant & Cell Physiology, 2003, 44(12): 1350-1358. |
| 48 | Zhang Z, Wei L, Zou X, et al. Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Annals of Botany, 2008, 102(4): 509-519. |
| 49 | Boualem A, Laporte P, Jovanovic M, et al. MicroRNA166 controls root and nodule development in Medicago truncatula. The Plant Journal: For Cell and Molecular Biology, 2008, 54(5): 876-887. |
| 50 | Moldovan D, Spriggs A, Yang J, et al. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. Journal of Experimental Botany, 2010, 61(1): 165-177. |
| 51 | Wang X R, Li W J, Chen B X, et al. Analysis ofmiRNA in water spinach (Ipomoea aquatica)under long-time high temperature. Acta Horticulturae Sinica, 2019, 46(3): 486-498. |
| 王杏茹, 李文静, 陈冰星, 等. 蕹菜耐受长时间高温后的miRNA分析. 园艺学报, 2019, 46(3): 486-498. | |
| 52 | Gutierrez L, Bussell J D, Pacurar D I, et al. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of auxin response factor transcripts and microRNA abundance. The Plant Cell, 2009, 21(10): 3119-3132. |
| 53 | Zhu Y Y, Zeng H Q, Dong C X, et al. microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L). Plant Science, 2010, 178(1): 23-29. |
| 54 | Zhang D F. A study of microRNA-mediated mechanisms of adventitious roots growth and antioxidant in submergenced maize seedlings. Baoding: Hebei Agricultural University, 2009. |
| 张丹凤. 淹水胁迫下microRNA介导的玉米不定根生长及抗氧化机制研究. 保定: 河北农业大学, 2009. | |
| 55 | Ma J, Zhao P, Liu S, et al. The control of developmental phase transitions by microRNAs and their targets in seed plants. International Journal of Molecular Sciences, 2020, 21(6): 1971. |
| 56 | Schwab R, Palatnik J F, Riester M, et al. Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 2005, 8(4): 517-527. |
| 57 | Yamasaki K, Kigawa T, Inoue M, et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of arabidopsis SBP-family transcription factors. Journal of Molecular Biology, 2004, 337(1): 49-63. |
| 58 | Stief A, Altmann S, Hoffmann K, et al. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell, 2014, 26(4): 1792-1807. |
| 59 | Xu X, Wang K, Pan J, et al. Small RNA sequencing identifies cucumber miRNA roles in waterlogging-triggered adventitious root primordia formation. Molecular Biology Reports, 2019, 46(6): 6381-6389. |
| 60 | Wang Y, Sun F, Cao H, et al. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One, 2012, 7(11): e48445. |
| 61 | Zhang K, Shi X, Zhao X, et al. Investigation of miR396 and growth-regulating factor regulatory network in maize grain filling. Acta Physiologiae Plantarum, 2015, 37(2): 1-12. |
| 62 | Gao P, Bai X, Yang L, et al. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta, 2010, 231(5): 991-1001. |
| 63 | Lin Y H, Tang L Q, Xu J, et al. Identification of genes and proteins involved in MAPK pathway response to submergence stress in soybean. Molecular Plant Breeding, 2020, 18(12): 3819-3824. |
| 林延慧, 唐力琼, 徐靖, 等. 大豆响应耐涝的MAPK信号通路相关基因及蛋白的鉴定分析. 分子植物育种, 2020, 18(12): 3819-3824. |
| [1] | 杨志民, 邢瑞, 丁鋆嘉, 庄黎丽. 基于转录组测序的高羊茅分蘖与株高相关差异表达基因分析[J]. 草业学报, 2022, 31(1): 145-163. |
| [2] | 周晶, 陈思齐, 史文娇, 阳伏林, 林辉, 林占熺. 巨菌草幼叶及根转录组功能基因测序及分析[J]. 草业学报, 2021, 30(2): 143-155. |
| [3] | 汪芳珍, 杨成行, 何子华, 林子茹, 曾浩源, 马清. 盐处理下旱生植物沙芥蛋白激酶相关基因的差异表达分析[J]. 草业学报, 2021, 30(10): 116-124. |
| [4] | 曾伟航, 程碧真, 彭燕, 李州. 甘露糖浸种对干旱胁迫下白三叶种子萌发及抗旱性的影响[J]. 草业学报, 2019, 28(7): 112-122. |
| [5] | 张旭, 聂刚, 黄琳凯, 唐露, 周洲, 刘福, 周洁, 邹静, 任思彦, 张新全. 植物生长调节剂对鸭茅种子产量的影响[J]. 草业学报, 2019, 28(6): 93-100. |
| [6] | 钱晨, 刘智微, 钟小仙, 吴娟子, 张建丽, 潘玉梅. 海滨雀稗自交结实突变体及野生型幼穗组织的转录组分析[J]. 草业学报, 2019, 28(5): 132-142. |
| [7] | 许蕾, 陈佩琳, 冯光燕, 钟旻依, 景婷婷, 黄琳凯, 张新全. 利用流式细胞仪鉴定鸭茅倍性[J]. 草业学报, 2019, 28(3): 74-84. |
| [8] | 唐露, 黄琳凯, 赵欣欣, 张旭, 聂刚, 张新全, 马啸. 四倍体鸭茅产量及其构成因素的QTL定位[J]. 草业学报, 2018, 27(11): 67-76. |
| [9] | 王新宇, 蒋林峰, 张新全, 黄琳凯, 李宁, 王鹏喜. 鸭茅DUS测试不同品种性状一致性分析[J]. 草业学报, 2016, 25(9): 104-116. |
| [10] | 黄梅芬, 薛世明, 高月娥, 李乔仙, 张美艳, 余梅, 钟声. 喜马拉雅鸭茅野生二倍体与同源四倍体农艺性状的对比研究[J]. 草业学报, 2016, 25(1): 207-216. |
| [11] | 蒋林峰,张新全,付玉凤,蒙芬,黄琳凯. 中国主要鸭茅品种农艺性状变异研究[J]. 草业学报, 2015, 24(3): 142-154. |
| [12] | 季杨,张新全,彭燕,梁小玉,黄琳凯,马啸,马迎梅. 干旱胁迫对鸭茅根、叶保护酶活性、渗透物质含量及膜质过氧化作用的影响[J]. 草业学报, 2014, 23(3): 144-151. |
| [13] | 蒋林峰,张新全,黄琳凯,马啸,严德飞,胡强,付玉凤. 鸭茅品种的SCoT遗传变异分析[J]. 草业学报, 2014, 23(1): 229-238. |
| [14] | 万刚,张新全,刘伟,谢文刚,周禾,彭燕. 鸭茅栽培品种与野生材料遗传多样性比较的SSR分析[J]. 草业学报, 2010, 19(6): 187-196. |
| [15] | 谢文刚, 张新全,陈永霞. 鸭茅杂交种的SSR分子标记鉴定及其遗传变异分析[J]. 草业学报, 2010, 19(2): 212-217. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||