

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2023, Vol. 32 ›› Issue (5): 147-158.DOI: 10.11686/cyxb2022183
• 研究论文 • 上一篇
收稿日期:2022-04-20
修回日期:2022-06-20
出版日期:2023-05-20
发布日期:2023-03-20
通讯作者:
赵桂琴
作者简介:E-mail: zhaogq@gsau.edu.cn基金资助:
Ji-kuan CHAI( ), Ze-liang JU, Gui-qin ZHAO(
), Ze-liang JU, Gui-qin ZHAO( )
)
Received:2022-04-20
Revised:2022-06-20
Online:2023-05-20
Published:2023-03-20
Contact:
Gui-qin ZHAO
摘要:
为了明确低温青贮中筛选出的优异耐低温乳酸菌菌株戊糖片球菌OL77在低温和低pH胁迫下的基因表达情况,探讨其耐低温机理,本研究通过同源克隆获得了OL77的CspP基因,运用GeNorm,NormFinder,BestKeeper和RefFinder程序,基于SYBR-Green的实时荧光定量PCR(RT-qPCR)筛选OL77在低温和低pH胁迫条件下表达稳定的内参基因,并利用其研究了CspP基因的低温表达模式。 结果表明:OL77在受到外界低温和低pH胁迫时表达较稳定的内参基因为Ldh、tufA和6PGDH,而GAPDH和GyrA是表达较不稳定的内参基因。采用同源克隆获得OL77的CspP基因片段,以Ldh和6PGDH为内参基因,对25、15和5 ℃下培养0、1、3、6、12、24 h的OL77中CspP基因的相对表达量进行分析,发现低温条件下CspP基因瞬时表达量激增,在5 ℃下上调86倍,这可能是OL77耐低温的主要原因,OL77对低温的适应性部分来自其较强的冷休克蛋白表达能力。CspP基因可用作OL77低温适应性的指示基因。
柴继宽, 琚泽亮, 赵桂琴. 低温和低pH胁迫下青贮乳酸菌OL77的内参基因筛选及CspP基因的表达模式分析[J]. 草业学报, 2023, 32(5): 147-158.
Ji-kuan CHAI, Ze-liang JU, Gui-qin ZHAO. Screening of internal reference genes of Lactobacillus strain OL77 and determination of CspP expression patterns under low-temperature and low-pH stress[J]. Acta Prataculturae Sinica, 2023, 32(5): 147-158.
基因名 Gene symbol | 引物序列(5′-3′正向/反向) Primer sequence (5′-3′,forward/reverse) | 产物长度 Product length (bp) | 扩增效率 Amplification efficiency (%) |
|---|---|---|---|
| tufA | GAGAAGCGTCACTATGCCCA/CACCAACTTGACGTGCCAAC | 166 | 101 |
| rpoD | GGTGTTGGTGGCTATCCCAA/GGTGTTGGTGGCTATCCCAA | 120 | 98 |
| recA | GGTGTTGGTGGCTATCCCAA/GGTGTTGGTGGCTATCCCAA | 114 | 96 |
| Ldh | TCGTCCTACACCGACAATGC/AGTAGCCGAAATGGCGCTTA | 151 | 99 |
| GyrA | ACCGGAATTGCTGTTGGAATG/CATCAGGTCGGCTGTGGTAG | 114 | 100 |
| GyrB | TCTACCCGGAAAGTTGGCAG/TGAAGCTTTCCCCACGTTCA | 166 | 98 |
| GAPDH | TTGAATTGACCATGAGCTGTATC/TCGGACGTATTGGTCGTTTA | 142 | 106 |
| 6PGDH | CTTCTTTGGCGATACAATTCG/CTAATGCGCCTAATTCACCA | 98 | 103 |
| CspP1 | TGTAATAGATTACCCCTTGT/TTCTTGTGATTCCCCAGATT | 164 | 99 |
| CspP2 | AACGACTGTTGCTTGTTCC/GCAAGTGTCAGGGTTTCTCT | 111 | 98 |
表1 候选内参基因及引物序列
Table 1 Candidate internal reference genes and primer sequences
基因名 Gene symbol | 引物序列(5′-3′正向/反向) Primer sequence (5′-3′,forward/reverse) | 产物长度 Product length (bp) | 扩增效率 Amplification efficiency (%) |
|---|---|---|---|
| tufA | GAGAAGCGTCACTATGCCCA/CACCAACTTGACGTGCCAAC | 166 | 101 |
| rpoD | GGTGTTGGTGGCTATCCCAA/GGTGTTGGTGGCTATCCCAA | 120 | 98 |
| recA | GGTGTTGGTGGCTATCCCAA/GGTGTTGGTGGCTATCCCAA | 114 | 96 |
| Ldh | TCGTCCTACACCGACAATGC/AGTAGCCGAAATGGCGCTTA | 151 | 99 |
| GyrA | ACCGGAATTGCTGTTGGAATG/CATCAGGTCGGCTGTGGTAG | 114 | 100 |
| GyrB | TCTACCCGGAAAGTTGGCAG/TGAAGCTTTCCCCACGTTCA | 166 | 98 |
| GAPDH | TTGAATTGACCATGAGCTGTATC/TCGGACGTATTGGTCGTTTA | 142 | 106 |
| 6PGDH | CTTCTTTGGCGATACAATTCG/CTAATGCGCCTAATTCACCA | 98 | 103 |
| CspP1 | TGTAATAGATTACCCCTTGT/TTCTTGTGATTCCCCAGATT | 164 | 99 |
| CspP2 | AACGACTGTTGCTTGTTCC/GCAAGTGTCAGGGTTTCTCT | 111 | 98 |
| 项目Item | tufA | rpoD | recA | Ldh | GyrA | GyrB | GAPDH | 6PGDH |
|---|---|---|---|---|---|---|---|---|
| pH | ||||||||
| 6.0 | 24.4 | 27.8 | 21.2 | 25.6 | 21.7 | 21.4 | 24.4 | 24.4 |
| 5.0 | 24.4 | 27.7 | 22.2 | 26.9 | 21.3 | 20.4 | 23.3 | 24.4 |
| 4.0 | 24.5 | 28.8 | 22.1 | 26.8 | 21.1 | 21.1 | 23.4 | 24.5 |
| 温度Temperature (℃) | ||||||||
| 25 | 24.7 | 27.3 | 22.3 | 24.4 | 20.5 | 20.8 | 21.3 | 24.7 |
| 15 | 24.8 | 24.5 | 22.1 | 24.9 | 21.1 | 21.0 | 20.7 | 24.8 |
| 5 | 24.6 | 22.0 | 22.5 | 24.8 | 20.5 | 21.3 | 21.9 | 24.6 |
| 均值Average | 24.6 | 26.4 | 22.1 | 25.6 | 21.0 | 21.0 | 22.5 | 24.6 |
| 标准差Standard deviation (SD) | 0.6 | 2.6 | 0.8 | 1.7 | 1.2 | 0.8 | 2.5 | 0.6 |
| 变异系数Coefficient of variation (CV, %) | 2.4 | 10.0 | 3.4 | 6.5 | 5.9 | 3.7 | 11.3 | 2.4 |
表2 8个候选内参基因的表达水平
Table 2 Expression levels of 8 candidate internal reference genes (Cq)
| 项目Item | tufA | rpoD | recA | Ldh | GyrA | GyrB | GAPDH | 6PGDH |
|---|---|---|---|---|---|---|---|---|
| pH | ||||||||
| 6.0 | 24.4 | 27.8 | 21.2 | 25.6 | 21.7 | 21.4 | 24.4 | 24.4 |
| 5.0 | 24.4 | 27.7 | 22.2 | 26.9 | 21.3 | 20.4 | 23.3 | 24.4 |
| 4.0 | 24.5 | 28.8 | 22.1 | 26.8 | 21.1 | 21.1 | 23.4 | 24.5 |
| 温度Temperature (℃) | ||||||||
| 25 | 24.7 | 27.3 | 22.3 | 24.4 | 20.5 | 20.8 | 21.3 | 24.7 |
| 15 | 24.8 | 24.5 | 22.1 | 24.9 | 21.1 | 21.0 | 20.7 | 24.8 |
| 5 | 24.6 | 22.0 | 22.5 | 24.8 | 20.5 | 21.3 | 21.9 | 24.6 |
| 均值Average | 24.6 | 26.4 | 22.1 | 25.6 | 21.0 | 21.0 | 22.5 | 24.6 |
| 标准差Standard deviation (SD) | 0.6 | 2.6 | 0.8 | 1.7 | 1.2 | 0.8 | 2.5 | 0.6 |
| 变异系数Coefficient of variation (CV, %) | 2.4 | 10.0 | 3.4 | 6.5 | 5.9 | 3.7 | 11.3 | 2.4 |
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(0.06) | tufA(0.13) | GyrA(0.23) | 6PGDH(0.33) | recA(0.54) | GAPDH(1.34) | GyrB(1.34) | rpoD(2.78) |
| 5.0 | GyrA(0.31) | rpoD(0.42) | GyrB(0.51) | Ldh(0.62) | 6PGDH(0.70) | tufA(1.14) | recA(1.53) | GAPDH(1.80) |
| 4.0 | Ldh(0.03) | GyrA(0.03) | tufA(0.08) | recA(0.48) | GyrB(1.11) | 6PGDH(1.30) | rpoD(1.69) | GAPDH(2.13) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(0.22) | 6PGDH(0.42) | Ldh(0.62) | rpoD(0.64) | GyrB(0.73) | recA(0.77) | GyrA(0.94) | GAPDH(1.57) |
| 15 | GyrB(0.11) | Ldh(0.69) | tufA(0.90) | recA(1.00) | 6PGDH(1.05) | GAPDH(1.43) | rpoD(1.67) | GyrA(1.88) |
| 5 | Ldh(0.10) | recA(0.43) | tufA(0.45) | 6PGDH(0.48) | GyrB(0.48) | rpoD(0.90) | GyrA(1.29) | GAPDH(2.32) |
| 总体Total | GyrB(0.26) | GyrA(0.34) | 6PGDH(0.57) | Ldh(0.60) | tufA(0.73) | GAPDH(0.73) | recA(0.87) | rpoD(0.92) |
表3 基于NormFinder分析候选内参基因的稳定性
Table 3 Stability analysis of candidate internal reference genes based on NormFinder
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(0.06) | tufA(0.13) | GyrA(0.23) | 6PGDH(0.33) | recA(0.54) | GAPDH(1.34) | GyrB(1.34) | rpoD(2.78) |
| 5.0 | GyrA(0.31) | rpoD(0.42) | GyrB(0.51) | Ldh(0.62) | 6PGDH(0.70) | tufA(1.14) | recA(1.53) | GAPDH(1.80) |
| 4.0 | Ldh(0.03) | GyrA(0.03) | tufA(0.08) | recA(0.48) | GyrB(1.11) | 6PGDH(1.30) | rpoD(1.69) | GAPDH(2.13) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(0.22) | 6PGDH(0.42) | Ldh(0.62) | rpoD(0.64) | GyrB(0.73) | recA(0.77) | GyrA(0.94) | GAPDH(1.57) |
| 15 | GyrB(0.11) | Ldh(0.69) | tufA(0.90) | recA(1.00) | 6PGDH(1.05) | GAPDH(1.43) | rpoD(1.67) | GyrA(1.88) |
| 5 | Ldh(0.10) | recA(0.43) | tufA(0.45) | 6PGDH(0.48) | GyrB(0.48) | rpoD(0.90) | GyrA(1.29) | GAPDH(2.32) |
| 总体Total | GyrB(0.26) | GyrA(0.34) | 6PGDH(0.57) | Ldh(0.60) | tufA(0.73) | GAPDH(0.73) | recA(0.87) | rpoD(0.92) |
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(0.28) | 6PGDH(0.35) | tufA(0.40) | GyrA(0.49) | recA(0.58) | GAPDH(1.04) | GyrB(1.44) | rpoD(2.89) |
| 5.0 | rpoD(0.32) | 6PGDH(0.42) | Ldh(0.50) | GyrA(0.52) | GyrB(0.65) | tufA(0.76) | recA(1.65) | GAPDH(1.91) |
| 4.0 | tufA(0.23) | Ldh(0.36) | GyrA(0.41) | recA(0.50) | 6PGDH(0.87) | rpoD(1.51) | GyrB(1.69) | GAPDH(2.34) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(0.37) | 6PGDH(0.50) | rpoD(0.67) | Ldh(0.74) | GyrB(0.87) | recA(0.98) | GAPDH(1.19) | GyrA(1.20) |
| 15 | GyrB(0.26) | tufA(0.53) | recA(0.63) | 6PGDH(0.75) | Ldh(0.92) | GAPDH(1.62) | rpoD(2.06) | GyrA(2.22) |
| 5 | Ldh(0.27) | recA(0.47) | tufA(0.56) | 6PGDH(0.58) | GyrB(0.62) | rpoD(0.97) | GyrA(1.22) | GAPDH(2.20) |
| 总体Total | tufA(0.46) | Ldh(0.60) | 6PGDH(0.62) | GyrB(0.95) | GyrA(1.24) | GAPDH(1.98) | recA(2.08) | rpoD(2.19) |
表4 候选内参基因的SD值
Table 4 SD value of candidate internal reference genes
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(0.28) | 6PGDH(0.35) | tufA(0.40) | GyrA(0.49) | recA(0.58) | GAPDH(1.04) | GyrB(1.44) | rpoD(2.89) |
| 5.0 | rpoD(0.32) | 6PGDH(0.42) | Ldh(0.50) | GyrA(0.52) | GyrB(0.65) | tufA(0.76) | recA(1.65) | GAPDH(1.91) |
| 4.0 | tufA(0.23) | Ldh(0.36) | GyrA(0.41) | recA(0.50) | 6PGDH(0.87) | rpoD(1.51) | GyrB(1.69) | GAPDH(2.34) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(0.37) | 6PGDH(0.50) | rpoD(0.67) | Ldh(0.74) | GyrB(0.87) | recA(0.98) | GAPDH(1.19) | GyrA(1.20) |
| 15 | GyrB(0.26) | tufA(0.53) | recA(0.63) | 6PGDH(0.75) | Ldh(0.92) | GAPDH(1.62) | rpoD(2.06) | GyrA(2.22) |
| 5 | Ldh(0.27) | recA(0.47) | tufA(0.56) | 6PGDH(0.58) | GyrB(0.62) | rpoD(0.97) | GyrA(1.22) | GAPDH(2.20) |
| 总体Total | tufA(0.46) | Ldh(0.60) | 6PGDH(0.62) | GyrB(0.95) | GyrA(1.24) | GAPDH(1.98) | recA(2.08) | rpoD(2.19) |
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(1.00) | tufA(2.21) | 6PGDH(2.83) | GyrA(3.22) | recA(5.00) | GAPDH(6.00) | GyrB(7.00) | rpoD(8.00) |
| 5.0 | rpoD(2.00) | GyrA(2.11) | Ldh(2.45) | 6PGDH(2.51) | GyrB(3.87) | tufA(6.00) | recA(7.00) | GAPDH(8.00) |
| 4.0 | Ldh(1.19) | tufA(2.06) | GyrA(2.06) | recA(4.00) | 6PGDH(5.48) | GyrB(5.69) | rpoD(6.74) | GAPDH(8.00) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(1.41) | 6PGDH(1.68) | Ldh(2.45) | rpoD(3.94) | recA(4.82) | GyrB(5.48) | GyrA(7.24) | GAPDH(7.74) |
| 15 | GyrB(1.32) | tufA(2.06) | Ldh(3.16) | 6PGDH(3.16) | recA(3.72) | GAPDH(6.00) | rpoD(7.00) | GyrA(8.00) |
| 5 | Ldh(1.00) | recA(1.68) | 6PGDH(3.46) | tufA(4.05) | GyrB(4.47) | rpoD(5.73) | GyrA(7.00) | GAPDH(8.00) |
| 总体Total | GyrB(2.00) | 6PGDH(2.06) | Ldh(2.21) | tufA(2.94) | GyrA(3.76) | GAPDH(6.00) | recA(7.00) | rpoD(8.00) |
表5 基于RefFinder分析候选内参基因的稳定性
Table 5 RefFinder based candidate internal reference gene stability
项目 Item | 排序Rank | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| pH | ||||||||
| 6.0 | Ldh(1.00) | tufA(2.21) | 6PGDH(2.83) | GyrA(3.22) | recA(5.00) | GAPDH(6.00) | GyrB(7.00) | rpoD(8.00) |
| 5.0 | rpoD(2.00) | GyrA(2.11) | Ldh(2.45) | 6PGDH(2.51) | GyrB(3.87) | tufA(6.00) | recA(7.00) | GAPDH(8.00) |
| 4.0 | Ldh(1.19) | tufA(2.06) | GyrA(2.06) | recA(4.00) | 6PGDH(5.48) | GyrB(5.69) | rpoD(6.74) | GAPDH(8.00) |
| 温度Temperature (℃) | ||||||||
| 25 | tufA(1.41) | 6PGDH(1.68) | Ldh(2.45) | rpoD(3.94) | recA(4.82) | GyrB(5.48) | GyrA(7.24) | GAPDH(7.74) |
| 15 | GyrB(1.32) | tufA(2.06) | Ldh(3.16) | 6PGDH(3.16) | recA(3.72) | GAPDH(6.00) | rpoD(7.00) | GyrA(8.00) |
| 5 | Ldh(1.00) | recA(1.68) | 6PGDH(3.46) | tufA(4.05) | GyrB(4.47) | rpoD(5.73) | GyrA(7.00) | GAPDH(8.00) |
| 总体Total | GyrB(2.00) | 6PGDH(2.06) | Ldh(2.21) | tufA(2.94) | GyrA(3.76) | GAPDH(6.00) | recA(7.00) | rpoD(8.00) |
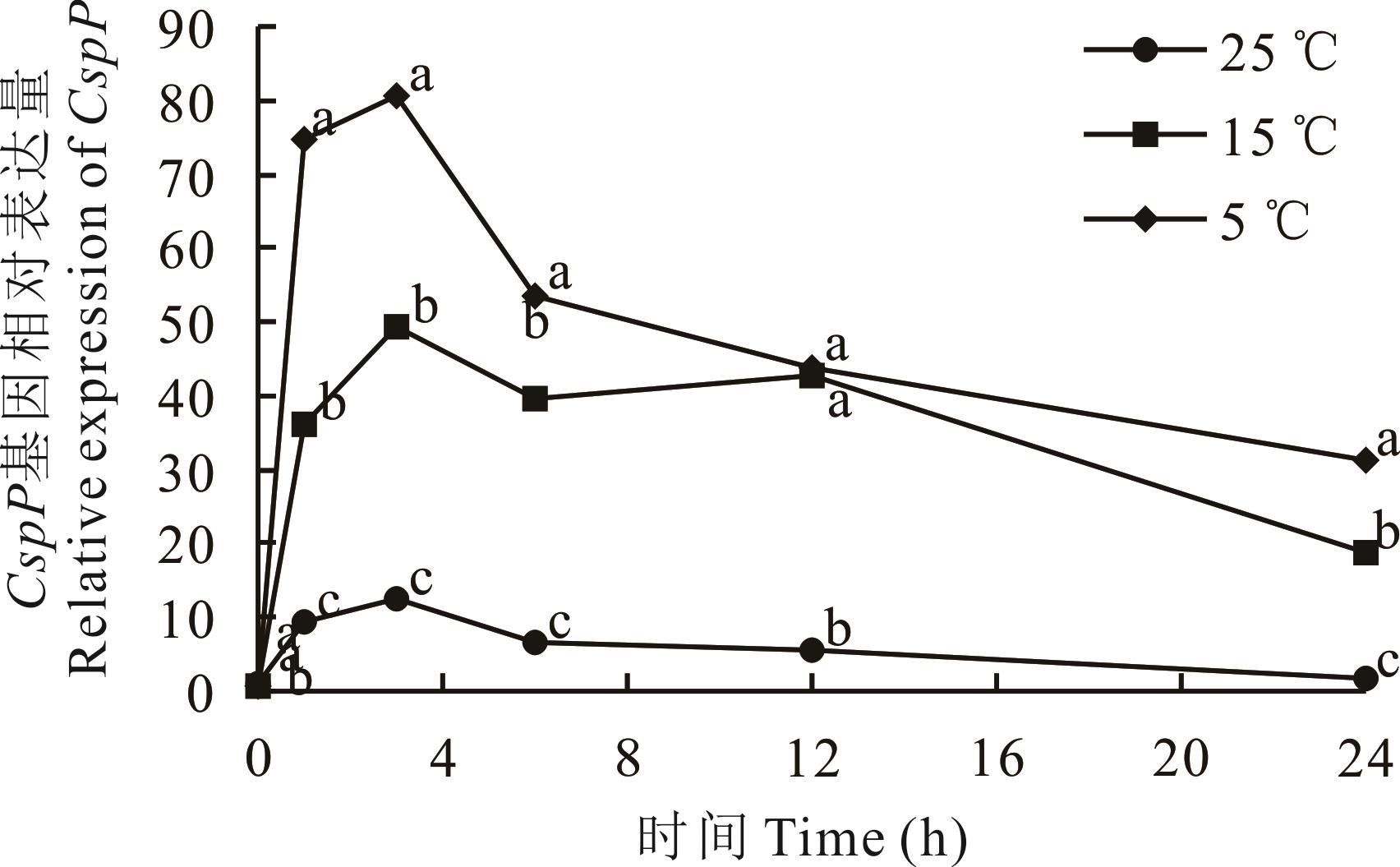
图5 不同温度下CspP基因的相对表达量同一时间不同字母表示不同温度处理间差异显著(P<0.05)。Different letters at the same time indicate significant differences among different temperature treatments (P<0.05).
Fig.5 The relative expression of CspP gene at different temperatures
| 1 | Derzelle S, Hallet B, Ferain T, et al. Improved adaptation to cold-shock, stationary-phase, and freezing stresses in Lactobacillus plantarum overproducing cold-shock proteins. Applied and Environmental Microbiology, 2003, 69(7): 4285-4290. |
| 2 | Fernandez M M L, De R H A P, De V G F. Survival rate and enzyme activities of Lactobacillus acidophilus following frozen storage. Cryobiology, 1998, 36(4): 315-319. |
| 3 | Saarela M, Virkajärvi I, Alakomi H L, et al. Influence of fermentation time, cryoprotectant and neutralization of cell concentrate on freeze-drying survival, storage stability, and acid and bile exposure of Bifidobacterium animalis ssp. lactis cells produced without milk-based ingredients. Journal of Applied Microbiology, 2005, 99(6): 1330-1339. |
| 4 | Wang Y, Corrieu G, Béal C. Fermentation pH and temperature influence the cryotolerance of Lactobacillus acidophilus RD758. Journal of Dairy Science, 2005, 88(1): 21-29. |
| 5 | Gachon C, Mingam A, Charrier B. Real-time PCR: What relevance to plant studies? Journal of Experimental Botany, 2004, 55(402): 1445-1454. |
| 6 | Udvardi M K, Czechowski T, Scheible W R. Eleven golden rules of quantitative RT-PCR. The Plant Cell, 2008, 20(7): 1736-1737. |
| 7 | Kozera B, Rapacz M. Reference genes in real-time PCR. Journal of Applied Genetics, 2013, 54(4): 391-406. |
| 8 | Pfaffl M W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 2001, 29(9): 45-46. |
| 9 | Jain M, Nijhawan A, Tyagi A, et al. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem and Biophysical Research Communication, 2006, 345(2): 646-651. |
| 10 | Dheda K, Huggett J F, Chang J S, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Analytical Biochemistry, 2005, 344(1): 141-143. |
| 11 | Jones P G, Inouye M. The cold-shock response-A hot topic. Molecular Microbiology, 1994, 11(4): 811-818. |
| 12 | Goldstein J, Pollitt N S, Inouye M. Major cold shock protein of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(1): 283-287. |
| 13 | Yamanaka K, Mitani T, Ogura T, et al. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Molecular Microbiology, 1994, 13(2): 301-312. |
| 14 | Hebraud M, Dubois E, Potier P, et al. Effect of growth temperatures on the protein levels in a psychrotrophic bacterium, Pseudomonas fragi. Journal of Bacteriology, 1994, 176(13): 4017-4024. |
| 15 | Willimsky G, Bang H, Fischer G, et al. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. Journal of Bacteriology, 1992, 174(20): 6326-6335. |
| 16 | Graumann P, Schröder K, Schmid R, et al. Cold shock stress-induced proteins in Bacillus subtilis. Journal of Bacteriology, 1996, 178(15): 4611-4619. |
| 17 | Derzelle S, Hallet B, Ferain T, et al. Cold shock induction of the cspL gene in Lactobacillus plantarum involves transcriptional regulation. Journal of Bacteriology, 2002, 184(19): 5518-5523. |
| 18 | Mayo B, Derzelle S, Fernández M, et al. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. Journal of Bacteriology, 1997, 179(9): 3039-3042. |
| 19 | Phadtare S. Recent developments in bacterial cold-shock response. Current Issues in Molecular Biology, 2004, 6(2): 125-136. |
| 20 | Wilkins J C, Homer K A, Beighton D. Altered protein expression of streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Applied and Environmental Microbiology, 2001, 67(8): 3396-3405. |
| 21 | Zhang L W, Liu C, Zhang Y H. Effect of over expression of cold shock protein on the fermentation properties of lactic acid bacteria. China Dairy Industry, 2018, 46(10): 15-18. |
| 张李伟, 刘畅, 张英华. 高效表达冷休克蛋白的乳酸菌发酵体系质构特性分析. 中国乳品工业, 2018, 46(10): 15-18. | |
| 22 | Yamamoto N, Aaano R, Yoshii H, et al. Archaeal community dynamics and detection of ammonia-oxidizing archaea during composting of cattle manure using culture-independent DNA analysis. Applied Microbiology & Biotechnology, 2011, 90(4): 1501-1510. |
| 23 | Zhou Y, Drouin P, Lafrenière C. Effect of temperature (5-25 ℃) on epiphytic lactic acid bacteria populations and fermentation of whole-plant corn silage. Journal of Applied Microbiology, 2016, 121(3): 657-671. |
| 24 | Zhang J, Guo G, Chen L, et al. Effect of applying lactic acid bacteria and propionic acid on fermentation quality and aerobic stability of oat-common vetch mixed silage on the Tibetan plateau. Animal Science Journal, 2015, 86: 595-602. |
| 25 | Zhang M, Lv H X, Tan Z F, et al. Improving the fermentation quality of wheat straw silage stored at low temperature by psychrotrophic lactic acid bacteria. Animal Science Journal, 2017, 88(2): 277-285. |
| 26 | Lin D D, Ju Z L, Chai J K, et al. Screening and identification of low temperature tolerant lactic acid bacterial epiphytes from oats on the Qinghai-Tibetan Plateau. Acta Prataculturae Sinica, 2022, 31(5): 103-114. |
| 蔺豆豆, 琚泽亮, 柴继宽, 等. 青藏高原燕麦附着耐低温乳酸菌的筛选与鉴定. 草业学报, 2022, 31(5): 103-114. | |
| 27 | Jarošová J, Kundu J K. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biology, 2010, 10(1): 146-155. |
| 28 | Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 2002, 3(7): 31-34. |
| 29 | Yang Z M, Chen Y, Hu B Y, et al. Identification and validation of reference genes for quantification of target gene expression with quantitative real-time PCR for tall fescue under four abiotic stresses. PLoS One, 2015, 10(3): 119-127. |
| 30 | Migocka M, Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Molecular Breeding, 2011, 28(3): 343-357. |
| 31 | Silver N, Best S, Jiang J, et al. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 2006, 7(1): 33-40. |
| 32 | Ruijter J M, Ramakers C, Hoogaars W M H, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research, 2009, 37(6): 45-55. |
| 33 | Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR date by geometric averaging of multiple internal control genes. Genome Biology, 2002, 3(7): 1-11. |
| 34 | Muck R E, Nadeau E M G, Mcallister T A, et al. Silage review: Recent advances and future uses of silage additives. Journal of Dairy Science, 2018, 101(5): 3980-4000. |
| 35 | Rapacz M, Stępień A, Skorupa K. Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): The effects of developmental stage and leaf age. Acta Physiologiae Plantarum, 2012, 34(5): 1723-1733. |
| 36 | Wrzesińska B, Kierzek R, Obrępalska-Stęplowska A. Evaluation of six commonly used reference genes for gene expression studies in herbicide-resistant Avena fatua biotypes. Weed Research, 2016, 56(4): 284-292. |
| 37 | Mallona I, Lischewski S, Weiss J, et al. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biology, 2010, 10(1): 4-12. |
| 38 | Han X J, Lu M Z, Chen Y C, et al. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS One, 2012, 7(8): 1-10. |
| 39 | Chao W S, Doğramaci M, Foley M E, et al. Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS One, 2012, 7(8): 42-53. |
| 40 | Lin X Z, He Z G, Li W X, et al. Validation of reference genes for real-time quantitative polymerase chain reaction analysis in Lactobacillus plantarum R23 under sulfur dioxide stress conditions. Australian Journal of Grape & Wine Research, 2018, 24(3): 390-395. |
| 41 | Trond L, Aparna S. Reference gene selection in Carnobacterium maltaromaticum, Lactobacillus curvatus, and Listeria innocua subjected to temperature and salt stress. Molecular Biotechnology, 2014, 56(3): 210-222. |
| 42 | Zhao W J, Li Y, Gao P F, et al. Validation of reference genes for real-time quantitative PCR studies in gene expression levels of Lactobacillus casei Zhang. Journal of Industrial Microbiology & Biotechnology, 2011, 38(9): 1279-1286. |
| 43 | Chen Y, Tan Z Q, Hu B Y, et al. Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiologia Plantarum, 2015, 155(2): 138-148. |
| 44 | Silveira E D, Alves-Ferreira M, Guimarães L A, et al. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology, 2009, 9(1): 84-93. |
| 45 | Robledo D, Hernández-Urcera J, Cal R M, et al. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics, 2014, 15(1): 643-648. |
| 46 | Bustin S A. Why the need for qPCR publication guidelines?-The case for MIQE. Methods, 2010, 50(4): 217-226. |
| 47 | Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods, 2010, 50(4): 227-230. |
| [1] | 李艳鹏, 魏娜, 翟庆妍, 李杭, 张吉宇, 刘文献. 全基因组水平白花草木樨TCP基因家族的鉴定及在干旱胁迫下表达模式分析[J]. 草业学报, 2023, 32(4): 101-111. |
| [2] | 孙守江, 唐艺涵, 马馼, 李曼莉, 毛培胜. 紫花苜蓿种子吸胀期胚根线粒体AsA-GSH循环对低温胁迫的响应[J]. 草业学报, 2023, 32(3): 152-162. |
| [3] | 刘晓婷, 姚拓. 高寒草地耐低温植物根际促生菌的筛选鉴定及特性研究[J]. 草业学报, 2022, 31(8): 178-187. |
| [4] | 蔺豆豆, 琚泽亮, 柴继宽, 赵桂琴. 青藏高原燕麦附着耐低温乳酸菌的筛选与鉴定[J]. 草业学报, 2022, 31(5): 103-114. |
| [5] | 刘亚男, 于人杰, 高燕丽, 康俊梅, 杨青川, 武志海, 王珍. 蒺藜苜蓿膜联蛋白MtANN2基因的表达模式及盐胁迫下的功能分析[J]. 草业学报, 2022, 31(5): 124-134. |
| [6] | 项洪涛, 郑殿峰, 何宁, 李琬, 王曼力, 王诗雅. 植物对低温胁迫的生理响应及外源脱落酸缓解胁迫效应的研究进展[J]. 草业学报, 2021, 30(1): 208-219. |
| [7] | 王玉萍, 郜春晓, 王盛祥, 何晓童. 低温弱光胁迫下芸豆叶片光抑制与类囊体膜脂构成变化[J]. 草业学报, 2020, 29(8): 116-125. |
| [8] | 张翔, 杨勇, 刘学勇, 向佐湘. 外源水杨酸对低温胁迫下海滨雀稗抗寒生理特征的影响[J]. 草业学报, 2020, 29(1): 117-124. |
| [9] | 项洪涛, 李琬, 何宁, 王雪扬, 郑殿峰, 王彤彤, 梁晓艳, 唐晓东, 李一丹. 苗期低温胁迫下烯效唑对红小豆根系抗寒生理及产量的影响[J]. 草业学报, 2019, 28(7): 92-102. |
| [10] | 王沛, 陈玖红, 王平, 马清, 田莉华, 陈有军, 周青平. 披碱草属植物抗逆性研究现状和存在的问题[J]. 草业学报, 2019, 28(5): 151-162. |
| [11] | 项洪涛, 齐德强, 李琬, 郑殿峰, 王月溪, 王彤彤, 王立志, 曾宪楠, 杨纯杰, 周行, 赵海东. 低温胁迫下外源ABA对开花期水稻叶鞘激素含量及抗寒生理的影响[J]. 草业学报, 2019, 28(4): 81-94. |
| [12] | 孙亚男, 林茹, 潘晓阳, 陈月, 陶磊, 郭长虹. 紫花苜蓿MsZAT10基因的克隆及其在烟草中的功能验证[J]. 草业学报, 2019, 28(12): 94-102. |
| [13] | 舒必超, 杨勇, 刘雪勇, 蒋元利, 向佐湘, 胡龙兴. 低温胁迫对狗牙根生理及基因表达的影响[J]. 草业学报, 2018, 27(11): 106-119. |
| [14] | 刘静, 陈振江, 李秀璋, 周景乐, 柳莉, 李春杰. 低温处理下外源水杨酸和脱落酸与内生真菌互作对醉马草共生体的影响[J]. 草业学报, 2018, 27(1): 142-151. |
| [15] | 张兰, 檀鹏辉, 滕珂, 闫蒙举, 何春燕, 甘露, 尹淑霞. 草地早熟禾荧光定量PCR分析中内参基因的筛选[J]. 草业学报, 2017, 26(3): 75-81. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||