

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2023, Vol. 32 ›› Issue (5): 190-202.DOI: 10.11686/cyxb2022207
• 综合评述 • 上一篇
收稿日期:2022-05-06
修回日期:2022-06-27
出版日期:2023-05-20
发布日期:2023-03-20
通讯作者:
伍国强
作者简介:E-mail: wugq08@126.com基金资助:
Xiao-han YANG( ), Guo-qiang WU(
), Guo-qiang WU( ), Ming WEI, Bei-chen WANG
), Ming WEI, Bei-chen WANG
Received:2022-05-06
Revised:2022-06-27
Online:2023-05-20
Published:2023-03-20
Contact:
Guo-qiang WU
摘要:
K+在植物生长发育及抵御非生物胁迫中扮演着重要角色。高亲和性K+转运蛋白(high-affinity K+ transporter, HKT)是最重要的阳离子转运蛋白家族之一,广泛参与植物K+和Na+吸收与转运。大量研究表明,HKT家族基因表达受Ca2+、腐植酸和胞嘧啶甲基化等调控;其介导植物体内Na+长距离运输以及维持K+和Mg2+稳态平衡,在植物抗逆性中发挥着关键作用。本研究对HKT家族的发现、结构与分类、生物学功能、表达与调控机制及其响应非生物胁迫等方面的研究成果加以综述,并对其未来研究方向进行展望,以期为农作物抗逆性遗传改良提供理论依据和基因资源。
杨小涵, 伍国强, 魏明, 王北辰. HKT在植物离子稳态和响应非生物逆境胁迫中的作用[J]. 草业学报, 2023, 32(5): 190-202.
Xiao-han YANG, Guo-qiang WU, Ming WEI, Bei-chen WANG. Function of high-affinity potassium transporters in maintaining ion homeostasis and other plant responses to abiotic stresses[J]. Acta Prataculturae Sinica, 2023, 32(5): 190-202.
物种 Species | 基因名称 Gene name | 登录号 Accession No. | 氨基酸数目 Amino acids (aa) | 分子量 MW (kDa) | 等电点 pI | 参考文献 References |
|---|---|---|---|---|---|---|
| 拟南芥A. thaliana | AtHKT1;1 | AAF68393 | 506 | 57.49 | 8.86 | [ |
| 节节麦A. tauschii | AetHKT2;4 | XP_020183163.1 | 508 | 55.94 | 8.96 | [ |
| 水稻O. sativa | OsHKT1;1 | CAD37183 | 552 | 61.86 | 8.95 | [ |
| OsHKT1;3 | CAD37185 | 531 | 59.30 | 9.41 | ||
| OsHKT1;4 | CAD37197 | 500 | 54.24 | 8.96 | ||
| OsHKT1;5 | BAB93392 | 554 | 60.22 | 8.79 | ||
| OsHKT2;1 | BAB61789 | 530 | 59.30 | 9.43 | ||
| OsHKT2;2 | BAB61791 | 530 | 59.15 | 9.49 | ||
| OsHKT2;3 | XP_015632753.1 | 509 | 56.37 | 9.13 | ||
| OsHKT2;4 | XP_015641899.1 | 509 | 56.12 | 8.74 | ||
| 毛果杨P. trichocarpa | PtHKT1;1 | XP_002325229 | 535 | 60.09 | 9.48 | [ |
| 番茄S. lycopersicum | SlHKT1;1 | NP_001289833.1 | 503 | 56.88 | 9.03 | [ |
| SlHKT1;2 | NP_001295273.1 | 555 | 63.42 | 9.11 | ||
| 高粱S. bicolor | SbHKT1;1 | KXG27071 | 582 | 63.74 | 9.71 | [ |
| SbHKT1;2 | XP_002457736 | 546 | 59.62 | 8.99 | ||
| SbHKT1;3 | XP_002451638 | 532 | 59.82 | 8.91 | ||
| SbHKT2;1 | XP_002438960 | 545 | 57.91 | 9.24 | ||
| 盐芥T. salsuginea | TsHKT1;1 | AFJ23835 | 505 | 57.27 | 8.78 | [ |
| TsHKT1;2 | BAJ34563 | 500 | 56.59 | 8.98 | ||
| 小麦T. aestivum | TaHKT1;5B1 | ABG33947 | 518 | 57.46 | 9.11 | [ |
| TaHKT1;5B2 | ABG33948 | 514 | 57.23 | 8.38 | ||
| TaHKT1;5D | ABG33945 | 516 | 57.30 | 8.90 | ||
| TaHKT2;1 | AAA52749 | 533 | 58.92 | 8.93 | ||
| TaHKT2;2 | AMB15006.1 | 508 | 56.01 | 9.04 | ||
| 一粒小麦T. monococcum | TmHKT1;4-A2 | ABK41857 | 554 | 60.59 | 9.75 | [ |
| TmHKT1;5-A | ABG33946 | 517 | 57.29 | 8.04 | ||
| TmHKT2;1-A1 | AMB15009.1 | 533 | 58.89 | 9.15 | ||
| 玉米Z. mays | ZmHKT1;5 | DAA54361 | 493 | 53.85 | 9.13 | [ |
| ZmHKT2;1 | XP_008645031 | 555 | 58.55 | 9.34 | ||
| 甜菜B. vulgaris | BvHKT1;1 | Bv8_198240_anxn | 280 | 31.95 | 9.39 | [ |
| BvHKT1;2 | Bv9_212780_gxzu | 505 | 57.13 | 9.12 | ||
| BvHKT1;3 | Bv9_212770_duhc | 511 | 57.85 | 9.42 |
表1 不同植物HKT基因
Table 1 The HKT genes in different plants
物种 Species | 基因名称 Gene name | 登录号 Accession No. | 氨基酸数目 Amino acids (aa) | 分子量 MW (kDa) | 等电点 pI | 参考文献 References |
|---|---|---|---|---|---|---|
| 拟南芥A. thaliana | AtHKT1;1 | AAF68393 | 506 | 57.49 | 8.86 | [ |
| 节节麦A. tauschii | AetHKT2;4 | XP_020183163.1 | 508 | 55.94 | 8.96 | [ |
| 水稻O. sativa | OsHKT1;1 | CAD37183 | 552 | 61.86 | 8.95 | [ |
| OsHKT1;3 | CAD37185 | 531 | 59.30 | 9.41 | ||
| OsHKT1;4 | CAD37197 | 500 | 54.24 | 8.96 | ||
| OsHKT1;5 | BAB93392 | 554 | 60.22 | 8.79 | ||
| OsHKT2;1 | BAB61789 | 530 | 59.30 | 9.43 | ||
| OsHKT2;2 | BAB61791 | 530 | 59.15 | 9.49 | ||
| OsHKT2;3 | XP_015632753.1 | 509 | 56.37 | 9.13 | ||
| OsHKT2;4 | XP_015641899.1 | 509 | 56.12 | 8.74 | ||
| 毛果杨P. trichocarpa | PtHKT1;1 | XP_002325229 | 535 | 60.09 | 9.48 | [ |
| 番茄S. lycopersicum | SlHKT1;1 | NP_001289833.1 | 503 | 56.88 | 9.03 | [ |
| SlHKT1;2 | NP_001295273.1 | 555 | 63.42 | 9.11 | ||
| 高粱S. bicolor | SbHKT1;1 | KXG27071 | 582 | 63.74 | 9.71 | [ |
| SbHKT1;2 | XP_002457736 | 546 | 59.62 | 8.99 | ||
| SbHKT1;3 | XP_002451638 | 532 | 59.82 | 8.91 | ||
| SbHKT2;1 | XP_002438960 | 545 | 57.91 | 9.24 | ||
| 盐芥T. salsuginea | TsHKT1;1 | AFJ23835 | 505 | 57.27 | 8.78 | [ |
| TsHKT1;2 | BAJ34563 | 500 | 56.59 | 8.98 | ||
| 小麦T. aestivum | TaHKT1;5B1 | ABG33947 | 518 | 57.46 | 9.11 | [ |
| TaHKT1;5B2 | ABG33948 | 514 | 57.23 | 8.38 | ||
| TaHKT1;5D | ABG33945 | 516 | 57.30 | 8.90 | ||
| TaHKT2;1 | AAA52749 | 533 | 58.92 | 8.93 | ||
| TaHKT2;2 | AMB15006.1 | 508 | 56.01 | 9.04 | ||
| 一粒小麦T. monococcum | TmHKT1;4-A2 | ABK41857 | 554 | 60.59 | 9.75 | [ |
| TmHKT1;5-A | ABG33946 | 517 | 57.29 | 8.04 | ||
| TmHKT2;1-A1 | AMB15009.1 | 533 | 58.89 | 9.15 | ||
| 玉米Z. mays | ZmHKT1;5 | DAA54361 | 493 | 53.85 | 9.13 | [ |
| ZmHKT2;1 | XP_008645031 | 555 | 58.55 | 9.34 | ||
| 甜菜B. vulgaris | BvHKT1;1 | Bv8_198240_anxn | 280 | 31.95 | 9.39 | [ |
| BvHKT1;2 | Bv9_212780_gxzu | 505 | 57.13 | 9.12 | ||
| BvHKT1;3 | Bv9_212770_duhc | 511 | 57.85 | 9.42 |

图1 HKT结构和分类(A) HKT蛋白的基本结构。HKT有8个跨膜结构域(M1A~M2D)和4个孔环结构域(PA~PD)。负责Na+/K+选择性的丝氨酸(绿色)和甘氨酸(粉红色)残基在第一个孔环区域中用箭头表示。NH2和COOH分别表示HKT蛋白的N端和C端。(B) HKT家族成员分为Ⅰ类和Ⅱ类两类。大多数Ⅰ类成员在选择性过滤器位置具有丝氨酸残基,而大多数Ⅱ类转运蛋白在相同位置含有甘氨酸残基。(A) Basic structure of the HKT proteins. HKT has eight transmembrane domains (M1A-M2D) and four pore-loop domains (PA-PD). Serine (green) and glycine (pink) residues, which are responsible for Na+/K+ selectivity, are indicated by arrows in the first pore-loop region. NH2 and COOH indicate N- and C-terminus of HKT protein, respectively. (B) HKT family members are divided into two classes, namely Class Ⅰ and Ⅱ. Most members of Class Ⅰ possess a serine residue in the selectivity filter position, whereas most Class Ⅱ transporters contain a glycine residue at the same position.
Fig.1 Structure and classification of HKT
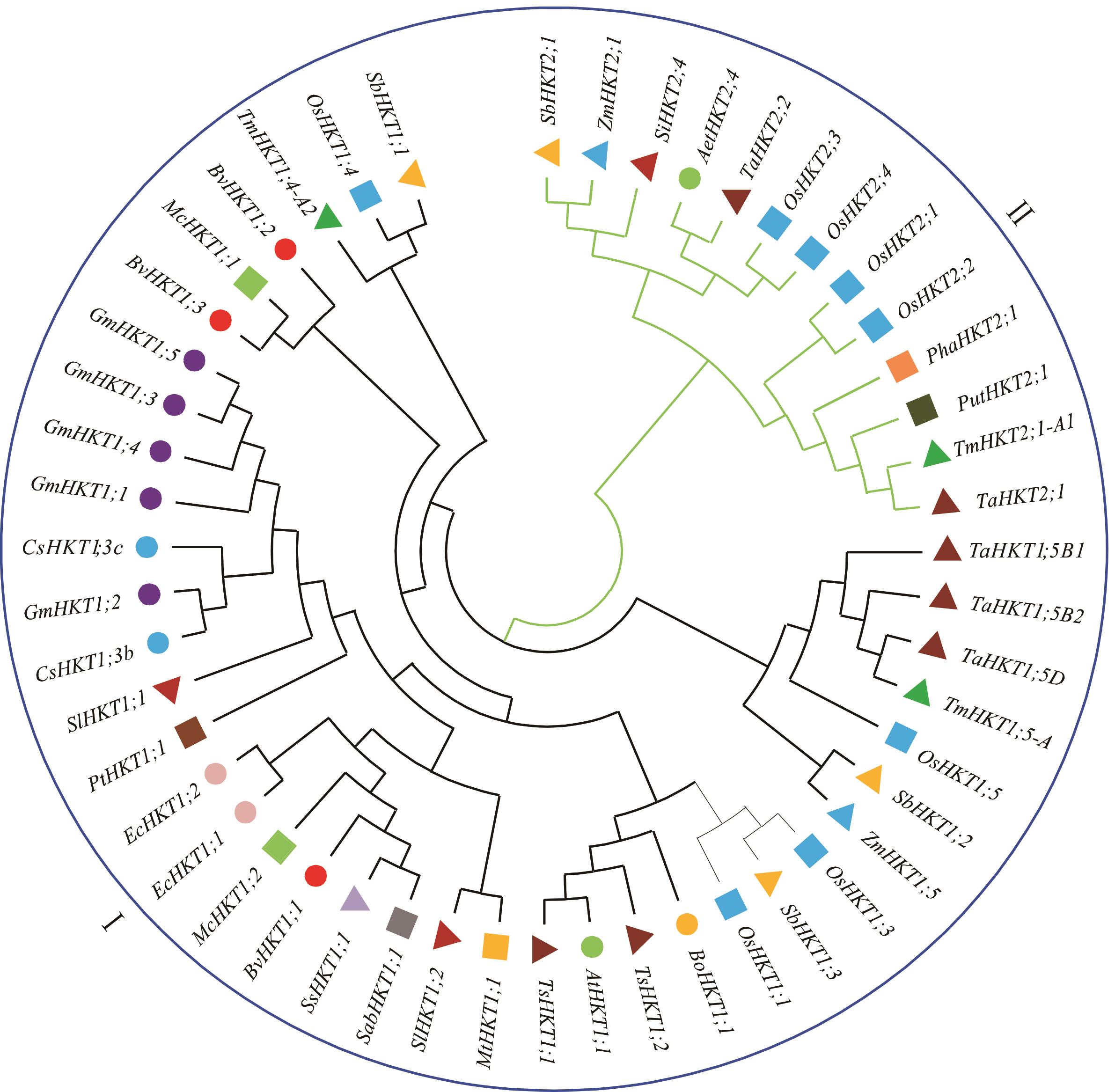
图2 植物HKT系统发育树HKT基因的来源、名称及蛋白登录号如下,其他物种、基因名称和登录号见表1。The source, name and accession number of the HKT gene are as follows, other source, name and accession number of HKT are shown in Table 1. 甘蓝Brassica oleracea: BoHKT1;1 (AFI81996); 甜橙Citrus sinensis: CsHKT1;3c (XP_006474312), CsHKT1;3b (XP_006474646); 赤桉Eucalyptus camaldulensis: EcHKT1;1 (AAF97728), EcHKT1;2 (AAD53890); 大豆Glycine max: GmHKT1;1 (KRH74148), GmHKT1;2 (KRH74149), GmHKT1;3 (XP_006582258), GmHKT1;4 (KRH49067), GmHKT1;5 (KRH25841); 蒺藜苜蓿Medicago truncatula: MtHKT1;1 (XP_003620952); 冰叶日中花M. crystallinum: McHKT1;1 (AAK52962), McHKT1;2 (AAO73474); 芦苇Phragmites australis: PhaHKT2;1 (BAE44385); 金银花Puccinellia tenuiflora: PutHKT2;1 (ACT21087); 大花海棠Salicornia bigelovii: SabHKT1;1 (ADG45565); 狗尾草Setaria italica: SiHKT2;4 (XP_004967240.1); 盐地碱蓬S. salsa: SsHKT1;1 (AAS20529).
Fig.2 Phylogenetic tree of the HKT in plants
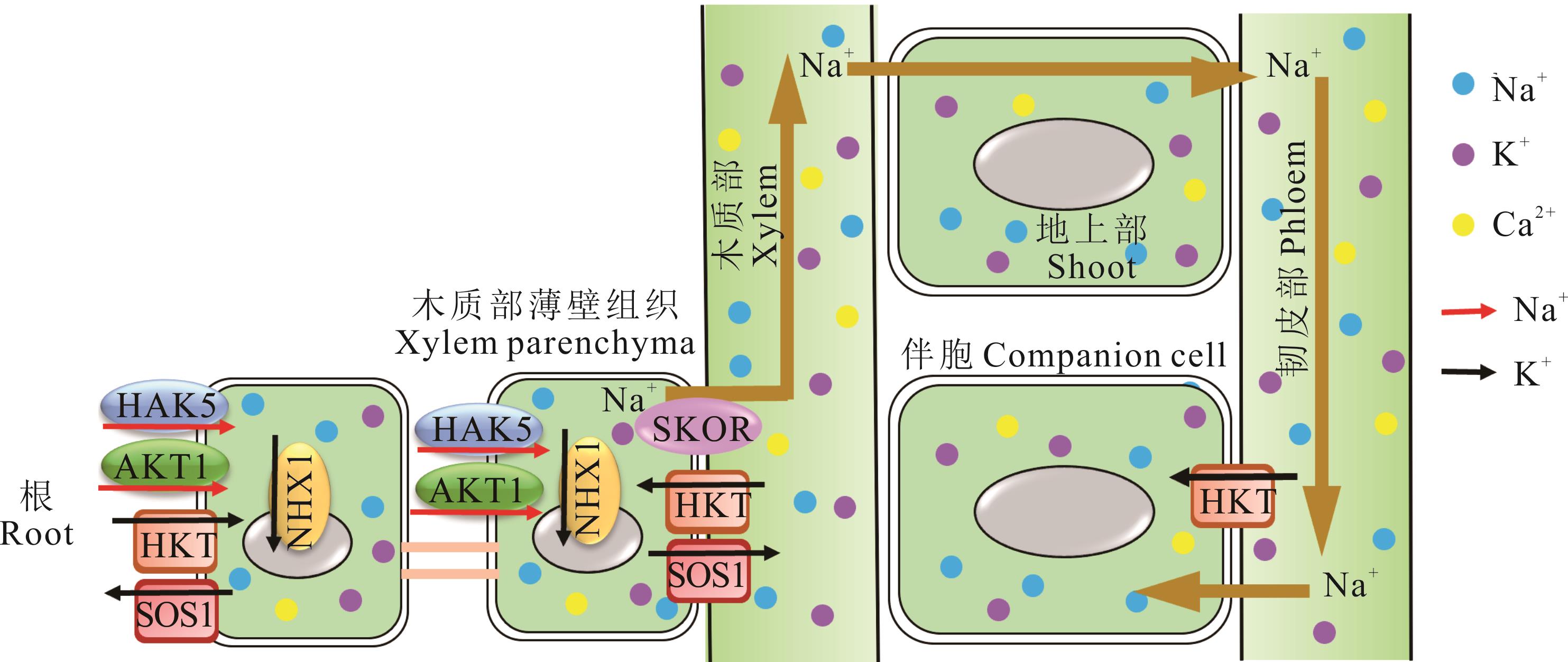
图3 盐胁迫下HKT维持Na+平衡高盐浓度的土壤允许Na+通过根部进入,再通过木质部在整个植物中运输,然后通过韧皮部再循环回根部。HKT通过将Na+从木质部输送到木质部薄壁细胞(流出)和从韧皮部输送到伴胞中来发挥作用,从而最大限度地减少因Na+积累而对植物造成的伤害。同时,SOS1与HKT互作维持Na+稳态。其中蓝色圆圈代表Na+,紫色圆圈代表K+,黄色圆圈代表Ca2+。红色箭头代表运输K+,黑色箭头代表运输Na+。High salt concentration in soil allows Na+ to enter through root and transport throughout the plant via the xylem vessel and is recirculated back to the root through phloem. HKT functions by transporting Na+ out of xylem vessel into xylem parenchyma (efflux) and out of phloem vessel into companion cell minimizing the harmful effects to the plant due to Na+ accumulation. Meanwhile, SOS1 cooperates with HKT to maintain Na+ homeostasis. Blue circles represent Na+, purple circles represent K+, and yellow circles represent Ca2+. Red arrows indicate transport K+, black arrows indicate transport Na+.
Fig.3 HKT maintains Na+ balance under salt stress
| 1 | Van O M J, Silletti S, Guida G, et al. A benzimidazole proton pump inhibitor increases growth and tolerance to salt stress in tomato. Frontiers in Plant Science, 2017, 8: 1220. |
| 2 | Zhang J L, Flowers T J, Wang S M. Differentiation of low-affinity Na+ uptake pathways and kinetics of the effects of K+ on Na+ uptake in the halophyte Suaeda maritima. Plant and Soil, 2012, 368(1/2): 629-640. |
| 3 | Wang Q, Guan C, Wang P, et al. The effect of AtHKT1;1 or AtSOS1 mutation on the expressions of Na+ or K+ transporter genes and ion homeostasis in Arabidopsis thaliana under salt stress. International Journal of Molecular Sciences, 2019, 20(5): 1085. |
| 4 | Mian A, Oomen R J, Isayenkov S, et al. Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. The Plant Journal, 2011, 68(3): 468-479. |
| 5 | Hazzouri K M, Khraiwesh B, Amiri K M A, et al. Mapping of HKT1;5 gene in barley using GWAS approach and its implication in salt tolerance mechanism. Frontiers in Plant Science, 2018, 9: 156. |
| 6 | Nieves-Cordones M, Martinez V, Benito B, et al. Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Frontiers in Plant Science, 2016, 7: 992. |
| 7 | Turcios A E, Papenbrock J, Tränkner M. Potassium, an important element to improve water use efficiency and growth parameters in quinoa (Chenopodium quinoa) under saline conditions. Journal of Agronomy and Crop Science, 2021, 207(4): 618-630. |
| 8 | Abdelaziz M E, Kim D, Ali S, et al. The endophytic fungus Piriformospora indica enhances Arabidopsis thaliana growth and modulates Na+/K+ homeostasis under salt stress conditions. Plant Science, 2017, 263: 107-115. |
| 9 | Song Z B, Wu X F, Gao Y L, et al. Genome-wide analysis of the HAK potassium transporter gene family reveals asymmetrical evolution in tobacco (Nicotiana tabacum). Genome, 2019, 62(4): 267-278. |
| 10 | Cao Y, Liang X, Yin P, et al. A domestication-associated reduction in K+-preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytologist, 2019, 222(1): 301-317. |
| 11 | Nieves-Cordones M, Lara A, Rodenas R, et al. Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell and Environment, 2019, 42(8): 2357-2371. |
| 12 | Dave A, Agarwal P, Agarwal P K. Mechanism of high affinity potassium transporter (HKT) towards improved crop productivity in saline agricultural lands. 3 Biotech, 2022, 12(2): 51. |
| 13 | Borkiewicz L, Polkowska-Kowalczyk L, Ciesla J, et al. Expression of maize calcium-dependent protein kinase (ZmCPK11) improves salt tolerance in transgenic Arabidopsis plants by regulating sodium and potassium homeostasis and stabilizing photosystem Ⅰ. Plant Physiology, 2020, 168(1): 38-57. |
| 14 | Huang S, Chen M, Zhao Y, et al. CBL4-CIPK5 pathway confers salt but not drought and chilling tolerance by regulating ion homeostasis. Environmental and Experimental Botany, 2020, 179: 104230. |
| 15 | Reguera E, Veatch J, Gedan K, et al. The effects of saltwater intrusion on germination success of standard and alternative crops. Environmental and Experimental Botany, 2020, 180: 104254. |
| 16 | Saleem M, Ji H, Amirullah A, et al. Pseudomonas syringae pv. tomato DC3000 growth in multiple gene knockouts predicts interactions among hormonal, biotic and abiotic stress responses. European Journal of Plant Pathology, 2017, 149(3): 779-786. |
| 17 | Saleem M, Law A D, Moe L A. Nicotiana roots recruit rare rhizosphere taxa as major root-inhabiting microbes. Microbial Ecology, 2016, 71(2): 469-472. |
| 18 | Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends in Plant Science, 2015, 20(4): 219-229. |
| 19 | Liu Z, Xie Q, Tang F, et al. The ThSOS3 gene improves the salt tolerance of transgenic Tamarix hispida and Arabidopsis thaliana. Frontiers in Plant Science, 2020, 11: 597480. |
| 20 | Platten J D, Cotsaftis O, Berthomieu P, et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends in Plant Science, 2006, 11(8): 372-374. |
| 21 | Pandey M, Penna S. Time course of physiological, biochemical, and gene expression changes under short-term salt stress in Brassica juncea L. The Crop Journal, 2017, 5(3): 219-230. |
| 22 | Dreyer I, Sussmilch F C, Fukushima K, et al. How to grow a tree: Plant voltage-dependent cation channels in the spotlight of evolution. Trends in Plant Science, 2021, 26(1): 41-52. |
| 23 | Garriga M, Raddatz N, Very A A, et al. Cloning and functional characterization of HKT1 and AKT1 genes of Fragaria spp.-Relationship to plant response to salt stress. Journal of Plant Physiology, 2017, 210: 9-17. |
| 24 | Mekawy A M M, Abdelaziz M N, Ueda A. Apigenin pretreatment enhances growth and salinity tolerance of rice seedlings. Plant Physiology and Biochemistry, 2018, 130: 94-104. |
| 25 | Rubio F, Gassmann W, Schroeder J I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science, 1995, 270(5242): 1660-1663. |
| 26 | Tada Y. The HKT transporter gene from Arabidopsis, AtHKT1;1, is dominantly expressed in shoot vascular tissue and root tips and is mild salt stress-responsive. Plants, 2019, 8(7): 204. |
| 27 | Nishijima R, Yoshida K, Motoi Y, et al. Genome-wide identification of novel genetic markers from RNA sequencing assembly of diverse Aegilops tauschii accessions. Molecular Genetics and Genomics, 2016, 291(4): 1681-1694. |
| 28 | Campbell M T, Bandillo N, Al Shiblawi F R A, et al. Allelic variants of OsHKT1;1 underlie the divergence between Indica and Japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genetics, 2017, 13(6): e1006823. |
| 29 | Xu M, Chen C, Cai H, et al. Overexpression of PeHKT1;1 improves salt tolerance in Populus. Genes, 2018, 9(10): 475. |
| 30 | Jaime-Perez N, Pineda B, Garcia-Sogo B, et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell and Environment, 2017, 40(5): 658-671. |
| 31 | Guo Q, Meng S, Tao S, et al. Overexpression of a samphire high-affinity potassium transporter gene SbHKT1 enhances salt tolerance in transgenic cotton. Acta Physiologiae Plantarum, 2020, 42(3): 36. |
| 32 | Ali Z, Park H C, Ali A, et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiology, 2012, 158(3): 1463-1474. |
| 33 | Xu B, Waters S, Byrt C S, et al. Structural variations in wheat HKT1;5 underpin differences in Na+ transport capacity. Cellular and Molecular Life Sciences, 2018, 75(6): 1133-1144. |
| 34 | Tounsi S, Saidi M N, Abdelhedi R, et al. Functional analysis of TmHKT1;4-A2 promoter through deletion analysis provides new insight into the regulatory mechanism underlying abiotic stress adaptation. Planta, 2021, 253(1): 18. |
| 35 | Jiang Z, Song G, Shan X, et al. Association analysis and identification of ZmHKT1;5 variation with salt-stress tolerance. Frontiers in Plant Science, 2018, 9: 1485. |
| 36 | Wu G Q, Liu Z X, Xie L L, et al. Genome-wide identification and expression analysis of the BvSnRK2 genes family in sugar beet (Beta vulgaris L.) under salt conditions. Journal of Plant Growth Regulation, 2021, 40(2): 519-532. |
| 37 | Gomez-Porras J L, Riano-Pachon D M, Benito B, et al. Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Frontiers in Plant Science, 2012, 3: 167. |
| 38 | Zhang S, Tong Y, Li Y, et al. Genome-wide identification of the HKT genes in five Rosaceae species and expression analysis of HKT genes in response to salt-stress in Fragaria vesca. Genes & Genomics, 2019, 41(3): 325-336. |
| 39 | Ariyarathna H A, Francki M G. Phylogenetic relationships and protein modelling revealed two distinct subfamilies of group Ⅰ HKT genes between crop and model grasses. Genome, 2016, 59(7): 509-517. |
| 40 | Imran S, Horie T, Katsuhara M. Expression and ion transport activity of rice OsHKT1;1 variants. Plants, 2019, 9(1): 16. |
| 41 | Riedelsberger J, Miller J K, Valdebenito-Maturana B, et al. Plant HKT channels: An updated view on structure, function and gene regulation. International Journal of Molecular Sciences, 2021, 22(4): 1892. |
| 42 | Ali A, Raddatz N, Pardo J M, et al. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Plant Physiology, 2021, 171(4): 546-558. |
| 43 | Hauser F, Horie T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell and Environment, 2010, 33(4): 552-565. |
| 44 | Diatloff E, Kumar R, Schachtman D P. Site directed mutagenesis reduces the Na+ affinity of HKT1, an Na+ energized high affinity K+ transporter. FEBS Letters, 1998, 432(1/2): 31-36. |
| 45 | Maser P, Hosoo Y, Goshima S, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(9): 6428-6433. |
| 46 | Almeida P M, de Boer G J, de Boer A H. Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SlHKT1;2 expressed in Xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Frontiers in Plant Science, 2014, 5: 600. |
| 47 | Romero-Aranda M R, Espinosa J, Gonzalez-Fernandez P, et al. Role of Na+ transporters HKT1;1 and HKT1;2 in tomato salt tolerance. I. Function loss of cheesmaniae alleles in roots and aerial parts. Plant Physiology and Biochemistry, 2021, 168: 282-293. |
| 48 | Pulipati S, Somasundaram S, Rana N, et al. Diversity of sodium transporter HKT1;5 in genus Oryza. Rice Science, 2022, 29(1): 31-46. |
| 49 | Munns R, James R A, Xu B, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology, 2012, 30(4): 360-364. |
| 50 | Byrt C S, Xu B, Krishnan M, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. The Plant Journal, 2014, 80(3): 516-526. |
| 51 | Prusty M R, Kim S R, Vinarao R, et al. Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Frontiers in Plant Science, 2018, 9: 417. |
| 52 | Henderson S W, Dunlevy J D, Wu Y, et al. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks. New Phytologist, 2018, 217(3): 1113-1127. |
| 53 | Wu Y, Henderson S W, Wege S, et al. The grapevine NaE sodium exclusion locus encodes sodium transporters with diverse transport properties and localisation. Journal of Plant Physiology, 2020, https://doi.org/10.1016/j.jplph.2020.153113. |
| 54 | Zhang M, Cao Y, Wang Z, et al. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytologist, 2018, 217(3): 1161-1176. |
| 55 | Kobayashi N I, Yamaji N, Yamamoto H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. The Plant Journal, 2017, 91(4): 657-670. |
| 56 | Shohan M U S, Sinha S, Nabila F H, et al. HKT1;5 transporter gene expression and association of amino acid substitutions with salt tolerance across rice genotypes. Frontiers in Plant Science, 2019, 10: 1420. |
| 57 | Davenport R J, Munoz-Mayor A, Jha D, et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell and Environment, 2007, 30(4): 497-507. |
| 58 | Kumar P, Kumar T, Singh S, et al. Potassium: A key modulator for cell homeostasis. Journal of Biotechnology, 2020, 324: 198-210. |
| 59 | Kronzucker H J, Britto D T. Sodium transport in plants: a critical review. New Phytologist, 2011, 189(1): 54-81. |
| 60 | Liu W, Schachtman D P, Zhang W. Partial deletion of a loop region in the high affinity K+ transporter HKT1 changes ionic permeability leading to increased salt tolerance. The Journal of Biological Chemistry, 2000, 275(36): 27924-27932. |
| 61 | Hmidi D, Messedi D, Corratgi-Faillie C, et al. Investigation of Na+ and K+ transport in halophytes: Functional analysis of the HmHKT2;1 transporter from Hordeum maritimum and expression under saline conditions. Plant and Cell Physiology, 2019, 60(11): 2423-2435. |
| 62 | Horie T, Brodsky D E, Costa A, et al. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiology, 2011, 156(3): 1493-1507. |
| 63 | Nowak M, Selmar D. Cellular distribution of alkaloids and their translocation via phloem and xylem: the importance of compartment pH. Plant Biology, 2016, 18(6): 879-882. |
| 64 | Hooymans J J M. Role of cell compartments in the redistribution of K and Na ions absorbed by the roots of intact barley plants. Zeitschrift für Pflanzenphysiologie, 1974, 73(3): 234-242. |
| 65 | Shen Q, Fu L, Dai F, et al. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genomics, 2016, 17(1): 889. |
| 66 | Horie T, Hauser F, Schroeder J I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends in Plant Science, 2009, 14(12): 660-668. |
| 67 | Shabala S, Shabala S, Cuin T A, et al. Xylem ionic relations and salinity tolerance in barley. The Plant Journal, 2010, 61(5): 839-853. |
| 68 | Vishwakarma K, Mishra M, Patil G, et al. Avenues of the membrane transport system in adaptation of plants to abiotic stresses. Critical Reviews in Biotechnology, 2019, 39(7): 861-883. |
| 69 | Carvalho M R, Losada J M, Niklas K J. Phloem networks in leaves. Current Opinion in Plant Biology, 2018, 43: 29-35. |
| 70 | Fu L, Shen Q, Kuang L, et al. Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiology and Biochemistry, 2018, 130: 248-257. |
| 71 | Quan R, Wang J, Hui J, et al. Improvement of salt tolerance using wild rice genes. Frontiers in Plant Science, 2017, 8: 2269. |
| 72 | Yamaguchi N, Ishikawa S, Abe T, et al. Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. Journal of Experimental Botany, 2012, 63(7): 2729-2737. |
| 73 | Babu N N, Krishnan S G, Vinod K K, et al. Marker aided incorporation of Saltol, a major QTL associated with seedling stage salt tolerance, into Oryza sativa ‘pusa basmati 1121’. Frontiers in Plant Science, 2017, 8: 41. |
| 74 | Dreyer I, Gomez-Porras J L, Riedelsberger J. The potassium battery: a mobile energy source for transport processes in plant vascular tissues. New Phytologist, 2017, 216(4): 1049-1053. |
| 75 | Sun J, Cao H, Cheng J, et al. Pumpkin CmHKT1;1 controls shoot Na+ accumulation via limiting Na+ transport from rootstock to scion in grafted cucumber. International Journal of Molecular Sciences, 2018, 19(9): 2648. |
| 76 | Houston K, Qiu J, Wege S, et al. Barley sodium content is regulated by natural variants of the Na+ transporter HvHKT1;5. Communications Biology, 2020, 3(1): 258. |
| 77 | Jabnoune M, Espeout S, Mieulet D, et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiology, 2009, 150(4): 1955-1971. |
| 78 | Tounsi S, Feki K, Saidi M N, et al. Promoter of the TmHKT1;4-A1 gene of Triticum monococcum directs stress inducible, developmental regulated and organ specific gene expression in transgenic Arabidopsis thaliana. World Journal of Microbiology and Biotechnology, 2018, 34(7): 99. |
| 79 | Pi H, Wendel B M, Helmann J D. Dysregulation of magnesium transport protects Bacillus subtilis against manganese and cobalt intoxication. Journal of Bacteriology, 2020, 202(7): e00711-19. |
| 80 | Lan W Z, Wang W, Wang S M, et al. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proceedings of the National Academy of Sciences, 2010, 107(15): 7089-7094. |
| 81 | Tang R J, Zhao F G, Garcia V J, et al. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proceedings of the National Academy of Sciences, 2015, 112(10): 3134-3139. |
| 82 | Zhang C, Li H, Wang J, et al. The rice high-affinity K+ transporter OsHKT2;4 mediates Mg2+ homeostasis under high-Mg2+ conditions in transgenic Arabidopsis. Frontiers in Plant Science, 2017, 8: 1823. |
| 83 | Rosas-Santiago P, Lagunas-Gomez D, Barkla B J, et al. Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. Journal of Experimental Botany, 2015, 66(9): 2733-2748. |
| 84 | Huang L, Kuang L, Wu L, et al. The HKT transporter HvHKT1;5 negatively regulates salt tolerance. Plant Physiology, 2020, 182(1): 584-596. |
| 85 | Ben Hsouna A, Ghneim-Herrera T, Ben Romdhane W, et al. Early effects of salt stress on the physiological and oxidative status of the halophyte Lobularia maritima. Functional Plant Biology, 2020, 47(10): 912-924. |
| 86 | Mahi H E, Perez-Hormaeche J, Luca A D, et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiology, 2019, 180(2): 1046-1065. |
| 87 | Munns R, Day D A, Fricke W, et al. Energy costs of salt tolerance in crop plants. New Phytologist, 2020, 225(3): 1072-1090. |
| 88 | Shabala S, Chen G, Chen Z H, et al. The energy cost of the tonoplast futile sodium leak. New Phytologist, 2020, 225(3): 1105-1110. |
| 89 | Shen Q, Fu L, Su T, et al. Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiology, 2020, 183(4): 1650-1662. |
| 90 | Choi W G, Hilleary R, Swanson S J, et al. Rapid, long-distance electrical and calcium signaling in plants. The Annual Review of Plant Biology, 2016, 67(1): 287-307. |
| 91 | Han Y, Yin S, Huang L, et al. A sodium transporter HvHKT1;1 confers salt tolerance in barley via regulating tissue and cell ion homeostasis. Plant and Cell Physiology, 2018, 59(10): 1976-1989. |
| 92 | Galon Y, Finkler A, Fromm H. Calcium-regulated transcription in plants. Molecular Plant, 2010, 3(4): 653-669. |
| 93 | Shkolnik D, Finkler A, Pasmanik-Chor M, et al. Calmodulin-binding transcription activator 6: A key regulator of Na+ homeostasis during germination. Plant Physiology, 2019, 180(2): 1101-1118. |
| 94 | Zhang X, Wang T, Liu M, et al. Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environmental and Experimental Botany, 2019, 157: 79-90. |
| 95 | Nardi S, Pizzeghello D, Muscolo A, et al. Physiological effects of humic substances on higher plants. Soil Biology and Biochemistry, 2002, 34(11): 1527-1536. |
| 96 | Türkmen Ö, Dursun A, Turan M, et al. Calcium and humic acid affect seed germination, growth, and nutrient content of tomato (Lycopersicon esculentum L.) seedlings under saline soil conditions. Acta Agriculturae Scandinavica, Section B-Soil & Plant Science, 2004, 54(3): 168-174. |
| 97 | Zhang W D, Wang P, Bao Z, et al. SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Frontiers in Plant Science, 2017, 8: 576. |
| 98 | Khaleda L, Park H J, Yun D J, et al. Humic acid confers high-affinity K+ transporter 1-mediated salinity stress tolerance in Arabidopsis. Molecules and Cells, 2017, 40(12): 966-975. |
| 99 | Cao Q, Feng Y, Dai X, et al. Dynamic changes of DNA methylation during wild strawberry (Fragaria nilgerrensis) tissue culture. Frontiers in Plant Science, 2021, 12: 765383. |
| 100 | Williams B P, Bechen L L, Pohlmann D A, et al. Somatic DNA demethylation generates tissue-specific methylation states and impacts flowering time. The Plant Cell, 2022, 34(4): 1189-1206. |
| 101 | Tounsi S, Feki K, Hmidi D, et al. Salt stress reveals differential physiological, biochemical and molecular responses in T. monococcum and T. durum wheat genotypes. Physiology and Molecular Biology of Plants, 2017, 23(3): 517-528. |
| 102 | Kumar S, Beena A S, Awana M, et al. Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA and Cell Biology, 2017, 36(4): 283-294. |
| 103 | Cha J Y, Kim T W, Choi J H, et al. Fungal laccase-catalyzed oxidation of naturally occurring phenols for enhanced germination and salt tolerance of Arabidopsis thaliana: A green route for synthesizing humic-like fertilizers. Journal of Agricultural and Food Chemistry, 2017, 65(6): 1167-1177. |
| 104 | Wang M, Qin L, Xie C, et al. Induced and constitutive DNA methylation in a salinity-tolerant wheat introgression line. Plant and Cell Physiology, 2014, 55(7): 1354-1365. |
| 105 | Li R, Zhu F, Duan D. Function analysis and stress-mediated cis-element identification in the promoter region of VqMYB15. Plant Signaling & Behavior, 2020, 15(7): 1773664. |
| 106 | Nawaz I, Iqbal M, Bliek M, et al. Salt and heavy metal tolerance and expression levels of candidate tolerance genes among four extremophile Cochlearia species with contrasting habitat preferences. Science of the Total Environment, 2017, 584/585: 731-741. |
| 107 | Nawaz I, Iqbal M, Hakvoort H W J, et al. Analysis of Arabidopsis thaliana HKT1 and Eutrema salsugineum/botschantzevii HKT1;2 promoters in response to salt stress in AtHKT1:1 mutant. Molecular Biotechnology, 2019, 61(6): 442-450. |
| 108 | Wu G Q, Wang J L, Li S J. Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes, 2019, 10(5): 401. |
| 109 | Bezouw V, Janssen E M, Ashrafuzzaman M, et al. Shoot sodium exclusion in salt stressed barley (Hordeum vulgare L.) is determined by allele specific increased expression of HKT1;5. Journal of Plant Physiology, 2019, 241: 153029. |
| 110 | Shkolnik-Inbar D, Adler G, Bar-Zvi D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. The Plant Journal, 2013, 73(6): 993-1005. |
| 111 | Wang R, Jing W, Xiao L, et al. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiology, 2015, 168(3): 1076-1090. |
| 112 | Hartmann F P, Tinturier E, Julien J L, et al. Between stress and response: Function and localization of mechanosensitive Ca2+ channels in herbaceous and perennial plants. International Journal of Molecular Sciences, 2021, 22(20): 11043. |
| 113 | Zhang H, Feng H, Zhang J, et al. Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. Journal of Experimental Botany, 2020, 71(14): 4345-4358. |
| 114 | Wang Y, Wu W H. Regulation of potassium transport and signaling in plants. Current Opinion in Plant Biology, 2017, 39: 123-128. |
| 115 | Cherel I, Gaillard I. The complex fine-tuning of K+ fluxes in plants in relation to osmotic and ionic abiotic stresses. International Journal of Molecular Sciences, 2019, 20(3): 715. |
| 116 | Sarah M M D S, Prado R D M, Souza J J P D, et al. Silicon supplied via foliar application and root to attenuate potassium deficiency in common bean plants. Scientific Reports, 2021, 11(1): 19690. |
| 117 | Tada Y, Endo C, Katsuhara M, et al. High-affinity K+ transporters from a halophyte, Sporobolus virginicus, mediate both K+ and Na+ transport in transgenic Arabidopsis, X. laevis oocytes and yeast. Plant and Cell Physiology, 2019, 60(1): 176-187. |
| 118 | Qiu L, Wu D, Ali S, et al. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theoretical and Applied Genetics, 2011, 122(4): 695-703. |
| 119 | Ferchichi S, Hessini K, Dell A E, et al. Hordeum vulgare and Hordeum maritimum respond to extended salinity stress displaying different temporal accumulation pattern of metabolites. Functional Plant Biology, 2018, 45(11): 1096-1109. |
| 120 | Liang W, Ma X, Wan P, et al. Plant salt-tolerance mechanism: A review. Biochemical and Biophysical Research Communications, 2018, 495(1): 286-291. |
| 121 | Hao S, Wang Y, Yan Y, et al. A review on plant responses to salt stress and their mechanisms of salt resistance. Horticulturae, 2021, 7(6): 132. |
| 122 | Tada Y, Ohnuma A. Comparative functional analysis of class Ⅱ potassium transporters, SvHKT2;1, SvHKT2;2, and HvHKT2;1, on ionic transport and salt tolerance in transgenic Arabidopsis. Plants, 2020, 9(6): 786. |
| 123 | Kawakami Y, Imran S, Katsuhara M, et al. Na+ transporter SvHKT1;1 from a halophytic turf grass is specifically upregulated by high Na+ concentration and regulates shoot Na+ concentration. International Journal of Molecular Sciences, 2020, 21(17): 6100. |
| 124 | Hou D, Zhao Z, Hu Q, et al. PeSNAC-1 a NAC transcription factor from moso bamboo (Phyllostachys edulis) confers tolerance to salinity and drought stress in transgenic rice. Tree Physiology, 2020, 40(12): 1792-1806. |
| 125 | Yang M, Lu K, Zhao F J, et al. Genome-wide association studies reveal the genetic basis of ionomic variation in rice. The Plant Cell, 2018, 30(11): 2720-2740. |
| 126 | Chen C, Travis A J, Hossain M, et al. Genome-wide association mapping of sodium and potassium concentration in rice grains and shoots under alternate wetting and drying and continuously flooded irrigation. Theoretical and Applied Genetics, 2021, 134(7): 2315-2334. |
| 127 | Chen N, Tong S, Tang H, et al. The PalERF109 transcription factor positively regulates salt tolerance via PalHKT1;2 in Populus alba var. pyramidalis. Tree Physiology, 2020, 40(6): 717-730. |
| 128 | Zhang T, Li Z Q, Wu G Q. Role of WRKY transcription factor in plant response to stresses. Biotechnology Bulletin, 2021, 37(10): 203-215. |
| 张桐, 李智强, 伍国强. WRKY转录因子在植物逆境响应中的作用. 生物技术通报, 2021, 37(10): 203-215. | |
| 129 | Zhu J K. Abiotic stress signaling and responses in plants. Cell, 2016, 167(2): 313-324. |
| 130 | Chen W J, Zhu T. Networks of transcription factors with roles in environmental stress response. Trends in Plant Science, 2004, 9(12): 591-596. |
| 131 | Yang C, Huang Y, Lv W, et al. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Science, 2020, 293: 110442. |
| 132 | Li Z, Liang F, Zhang T, et al. Enhanced tolerance to drought stress resulting from Caragana korshinskii CkWRKY33 in transgenic Arabidopsis thaliana. BMC Genomic Data, 2021, 22(1): 11. |
| 133 | Li J, Han Y, Liu L, et al. qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. Journal of Experimental Botany, 2015, 66(9): 2723-2732. |
| 134 | Li P, Pan T, Wang H, et al. Natural variation of ZmHKT1 affects root morphology in maize at the seedling stage. Planta, 2019, 249(3): 879-889. |
| [1] | 王园, 王晶, 李淑霞. 紫花苜蓿MsBBX24基因的克隆及耐盐性分析[J]. 草业学报, 2023, 32(3): 107-117. |
| [2] | 苗涵, 魏莱, 杨燕萍, 车永和. 海水胁迫下冰草幼苗期耐盐性指标筛选[J]. 草业学报, 2023, 32(3): 200-211. |
| [3] | 何海锋, 吴娜, 刘吉利, 许兴. 盐碱条件下施磷对柳枝稷生长发育及耐盐性的影响[J]. 草业学报, 2022, 31(10): 64-74. |
| [4] | 王晔, 陈慧萍, 李润枝, 彭真, 范希峰, 武菊英, 段留生. 奇岗微繁技术建立及幼苗耐盐性评价[J]. 草业学报, 2021, 30(6): 214-220. |
| [5] | 陈雅琦, 苏楷淇, 陈泰祥, 李春杰. 混合盐碱胁迫对醉马草种子萌发及幼苗生理特性的影响[J]. 草业学报, 2021, 30(3): 137-157. |
| [6] | 李倩, 李晓霞, 程丽琴, 陈双燕, 齐冬梅, 杨伟光, 高利军, 新巴音, 刘公社. 羊草LcCBF6基因的表达特性和功能研究[J]. 草业学报, 2021, 30(10): 105-115. |
| [7] | 熊雪, 桂维阳, 刘沫含, 陈继辉, 张英俊. 不同紫花苜蓿品种在均匀与不均匀盐胁迫下的耐盐性评价[J]. 草业学报, 2018, 27(9): 67-76. |
| [8] | 柯丹霞, 彭昆鹏, 夏远君, 朱玉莹, 张丹丹. 盐胁迫应答基因GmWRKY6的克隆及转基因百脉根的抗盐分析[J]. 草业学报, 2018, 27(8): 95-106. |
| [9] | 麻冬梅, 秦楚. AtSOS基因在紫花苜蓿中的表达及其耐盐性研究[J]. 草业学报, 2018, 27(6): 81-91. |
| [10] | 才华, 孙娜, 宋婷婷. 人工改造野生大豆GsDREB2基因对植物耐盐和耐渗透胁迫能力的影响[J]. 草业学报, 2018, 27(6): 168-176. |
| [11] | 柴艳, 孙宗玖, 李培英, 巴德木其其格, 张向向, 杨静. 新疆狗牙根种质芽期耐盐性综合评价[J]. 草业学报, 2017, 26(8): 154-167. |
| [12] | 伍国强, 冯瑞军, 李善家, 王春梅, 焦琦, 刘海龙. 盐处理对甜菜生长和渗透调节物质积累的影响[J]. 草业学报, 2017, 26(4): 169-177. |
| [13] | 麻冬梅, 秦楚, 倪星, 许兴, 郭伶娜. 高羊茅应答盐胁迫的AtSOS途径基因的效应分析[J]. 草业学报, 2016, 25(12): 170-179. |
| [14] | 贾新平,邓衍明,孙晓波,梁丽建. 盐胁迫对海滨雀稗生长和生理特性的影响[J]. 草业学报, 2015, 24(12): 204-212. |
| [15] | 张金林,李惠茹,郭姝媛,王锁民,施华中,韩庆庆,包爱科,马清. 高等植物适应盐逆境研究进展[J]. 草业学报, 2015, 24(12): 220-236. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||