

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (1): 61-74.DOI: 10.11686/cyxb2023076
胡尚钦1,2( ), 汪军成1,3, 姚立蓉1,3, 司二静1,3, 马小乐1,3, 杨轲1,3, 张宏1,3, 孟亚雄1,3, 王化俊1,3, 李葆春1,2(
), 汪军成1,3, 姚立蓉1,3, 司二静1,3, 马小乐1,3, 杨轲1,3, 张宏1,3, 孟亚雄1,3, 王化俊1,3, 李葆春1,2( )
)
收稿日期:2023-03-13
修回日期:2023-05-15
出版日期:2024-01-20
发布日期:2023-11-23
通讯作者:
李葆春
作者简介:E-mail: libc@gsau.edu.cn基金资助:
Shang-qin HU1,2( ), Jun-cheng WANG1,3, Li-rong YAO1,3, Er-jing SI1,3, Xiao-le MA1,3, Ke YANG1,3, Hong ZHANG1,3, Ya-xiong MENG1,3, Hua-jun WANG1,3, Bao-chun LI1,2(
), Jun-cheng WANG1,3, Li-rong YAO1,3, Er-jing SI1,3, Xiao-le MA1,3, Ke YANG1,3, Hong ZHANG1,3, Ya-xiong MENG1,3, Hua-jun WANG1,3, Bao-chun LI1,2( )
)
Received:2023-03-13
Revised:2023-05-15
Online:2024-01-20
Published:2023-11-23
Contact:
Bao-chun LI
摘要:
醛酮还原酶(AKR)是构成Shaker型K+通道蛋白的保守核心结构域,在植物应对非生物胁迫时起到关键作用。本研究采用西北旱区典型盐生植物盐生草作为研究材料,基于课题组前期盐生草根系盐胁迫转录组学数据分析结果,筛选并克隆得到耐盐基因HgAKR6C。HgAKR6C基因蛋白质编码区(CDS)全长951 bp,共编码氨基酸317个。系统发育进化树分析表明,HgAKR6C与拟南芥中AtAKR6C1基因亲缘关系最近。亚细胞定位表明该基因可能主要定位于细胞质和细胞核中。qRT-PCR结果表明HgAKR6C在盐处理24 h时表达量达到峰值。构建酵母异源表达载体转化缺陷型菌株发现,HgAKR6C基因可能参与Na+的外排和介导K+的吸收。综上所述,HgAKR6C具有调节盐生草耐盐性的功能,而盐生草根系耐盐基因HgAKR6C的调控机制还需进一步的研究验证。
胡尚钦, 汪军成, 姚立蓉, 司二静, 马小乐, 杨轲, 张宏, 孟亚雄, 王化俊, 李葆春. 盐生草根系基因HgAKR6C的克隆与初步功能分析[J]. 草业学报, 2024, 33(1): 61-74.
Shang-qin HU, Jun-cheng WANG, Li-rong YAO, Er-jing SI, Xiao-le MA, Ke YANG, Hong ZHANG, Ya-xiong MENG, Hua-jun WANG, Bao-chun LI. Cloning and preliminary functional analysis of the root gene HgAKR6C of Halogeton glomeratus[J]. Acta Prataculturae Sinica, 2024, 33(1): 61-74.
引物名称 Name of primers | 引物序列 Sequences of primers (5′→3′) |
|---|---|
| HgAKR6C-F1 | GC |
| HgAKR6C-R1 | TC |
| HgAKR6C-F2 | CC |
| HgAKR6C-R2 | GC |
| HgAKR6C-q1F | AGGAAGCACATCGTTGAGGG |
| HgAKR6C-q1R | TTCATTGCCCGGACAGTCTC |
| HgActin-F | TGTTCTCAGTGGTGGTACAA |
| HgActin-R | GTGCCACCACCTTAATCTTC |
表 1 试验所用引物
Table 1 Primers used in the study
引物名称 Name of primers | 引物序列 Sequences of primers (5′→3′) |
|---|---|
| HgAKR6C-F1 | GC |
| HgAKR6C-R1 | TC |
| HgAKR6C-F2 | CC |
| HgAKR6C-R2 | GC |
| HgAKR6C-q1F | AGGAAGCACATCGTTGAGGG |
| HgAKR6C-q1R | TTCATTGCCCGGACAGTCTC |
| HgActin-F | TGTTCTCAGTGGTGGTACAA |
| HgActin-R | GTGCCACCACCTTAATCTTC |

图1 HgAKR6C基因克隆PCR验证M: D15000+2000 DNA分子量标准D15000+2000 DNA marker; 1~4: 扩增HgAKR6C的片段Amplified HgAKR6C fragments. a用于后续构建亚细胞定位表达载体,b用于后续构建酵母异源表达载体。a was used for the subsequent construction of subcellular localization expression vectors, and b was used for the subsequent construction of yeast heterologous expression vectors.
Fig.1 The PCR validation of HgAKR6C
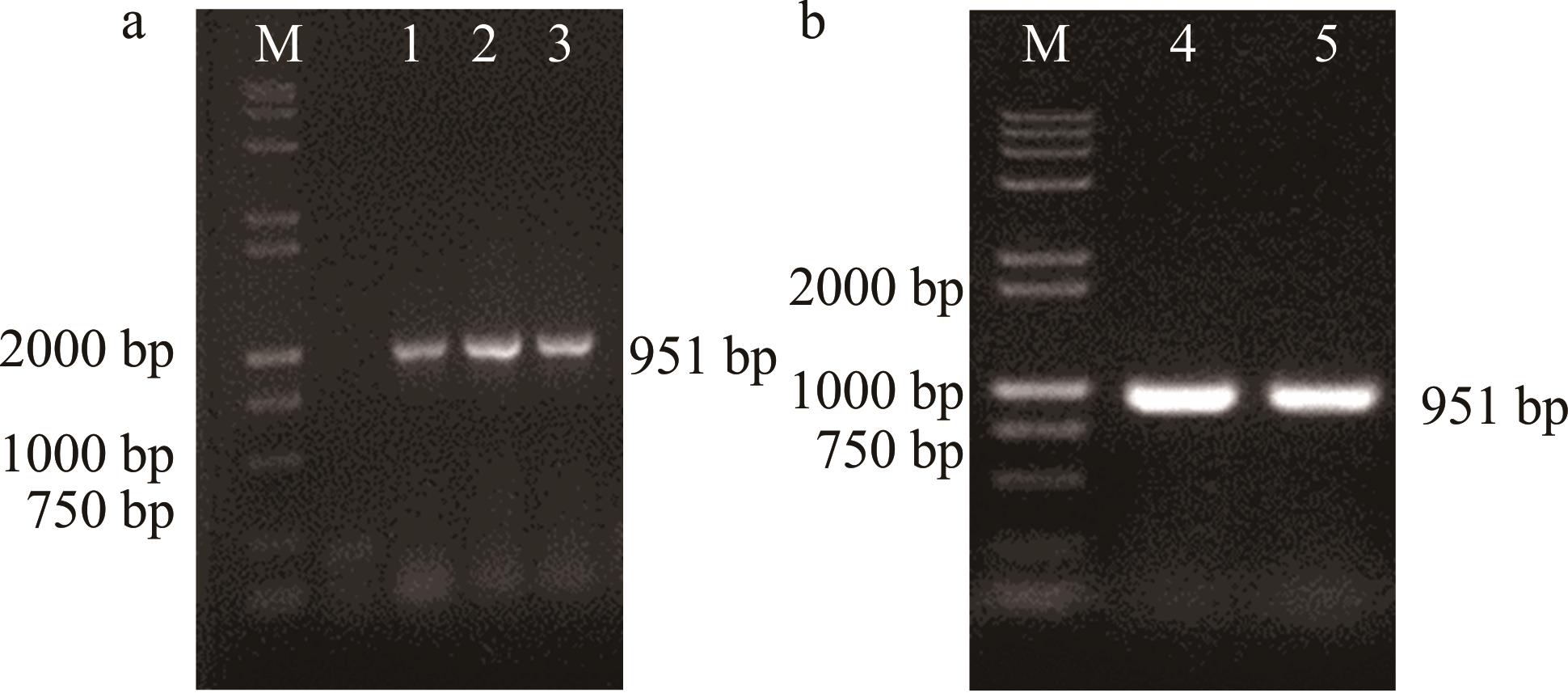
图2 重组载体菌液PCR验证1~5: 扩增重组载体的片段The fragments of amplified recombinant vectors. a为重组亚细胞定位表达载体,b为重组酵母异源表达载体。a is the recombinant subcellular locational expression vector, and b is the recombinant yeast heterologous expression vector.
Fig.2 PCR verification of recombinant vector solution
| 理化性质Physical and chemical properties | 预测结果Prediction results |
|---|---|
| 编码的氨基酸数Number of amino acids | 317 |
| 理论等电点Theoretical pI | 6.25 |
| 蛋白质分子量Molecular weight (Da) | 35151.21 |
| 分子式Formula | C1575H2473N419O468S12 |
| 负电荷的残基总数(天冬氨酸+谷氨酸)Total number of negatively charged residues (Asparticacid+glutamicacid, Asp+Glu) | 36 |
| 正电荷的残基总数(精氨酸+赖氨酸)Total number of positively charged residues (Arginine+lysine, Arg+Lys) | 34 |
| 脂肪系数Aliphatic index | 90.41 |
| 亲水性平均值Grand average of hydropathicity | -0.219 |
| 不稳定系数The instability index (II) | 32.06 |
表2 HgAKR6C基因的理化性质分析
Table 2 Analysis of physical and chemical properties of HgAKR6C
| 理化性质Physical and chemical properties | 预测结果Prediction results |
|---|---|
| 编码的氨基酸数Number of amino acids | 317 |
| 理论等电点Theoretical pI | 6.25 |
| 蛋白质分子量Molecular weight (Da) | 35151.21 |
| 分子式Formula | C1575H2473N419O468S12 |
| 负电荷的残基总数(天冬氨酸+谷氨酸)Total number of negatively charged residues (Asparticacid+glutamicacid, Asp+Glu) | 36 |
| 正电荷的残基总数(精氨酸+赖氨酸)Total number of positively charged residues (Arginine+lysine, Arg+Lys) | 34 |
| 脂肪系数Aliphatic index | 90.41 |
| 亲水性平均值Grand average of hydropathicity | -0.219 |
| 不稳定系数The instability index (II) | 32.06 |

图4 HgAKR6C基因的生物信息学分析预测a: 基因编码蛋白的跨膜区预测Prediction map of transmembrane region of protein encoded by gene; b: 信号肽预测Signal peptide prediction; c: 基因编码蛋白的亲疏水性分析Hydrophobicity analysis of protein encoded by gene; d: 基因编码蛋白的磷酸化分析Phosphorylation analysis of protein encoded by gene; e: 结构域预测Domain prediction.
Fig.4 Bioinformatics analysis and prediction of HgAKR6C genes
| 定位Positioning | 预测占比Forecast proportion (%) |
|---|---|
| 细胞质Cytoplasm | 60.9 |
| 细胞核 | 13.0 |
| 线粒体Mitochondrium | 13.0 |
| 细胞骨架Cytoskeleton | 4.3 |
表3 亚细胞定位预测
Table 3 Subcellular localization prediction (%)
| 定位Positioning | 预测占比Forecast proportion (%) |
|---|---|
| 细胞质Cytoplasm | 60.9 |
| 细胞核 | 13.0 |
| 线粒体Mitochondrium | 13.0 |
| 细胞骨架Cytoskeleton | 4.3 |

图 5 HgAKR6C同源蛋白序列比对Hg: 盐生草H. glomeratus; Md: 苹果Malus domestica; Ag: 旱芹Apium graveolens; Gm: 大豆Glycine max; Ms: 苜蓿Medicago sativa; Ge: 刺甘草Glycyrrhiza echinata; Gg: 洋甘草Glycyrrhiza glabra; Sr: 长喙田菁Sesbania rostrata; Ps: 罂粟Papaver somniferum; Fa: 草莓Fragaria ananassa; Zm: 玉米Zea mays; Os: 水稻O. sativa; Hv: 大麦Hordeum vulgare; Ta: 小麦Triticum aestivum; 无芒雀麦Bromus inermis; Af: 野燕麦Avena fatua; Xv: 翡若翠科黑炭木属Xerophyta viscosa; Dp: 毛地黄Digitalis purpurea; At: 拟南芥A. thaliana. 下同The same below.
Fig.5 Sequence alignment of HgAKR6C homologous protein
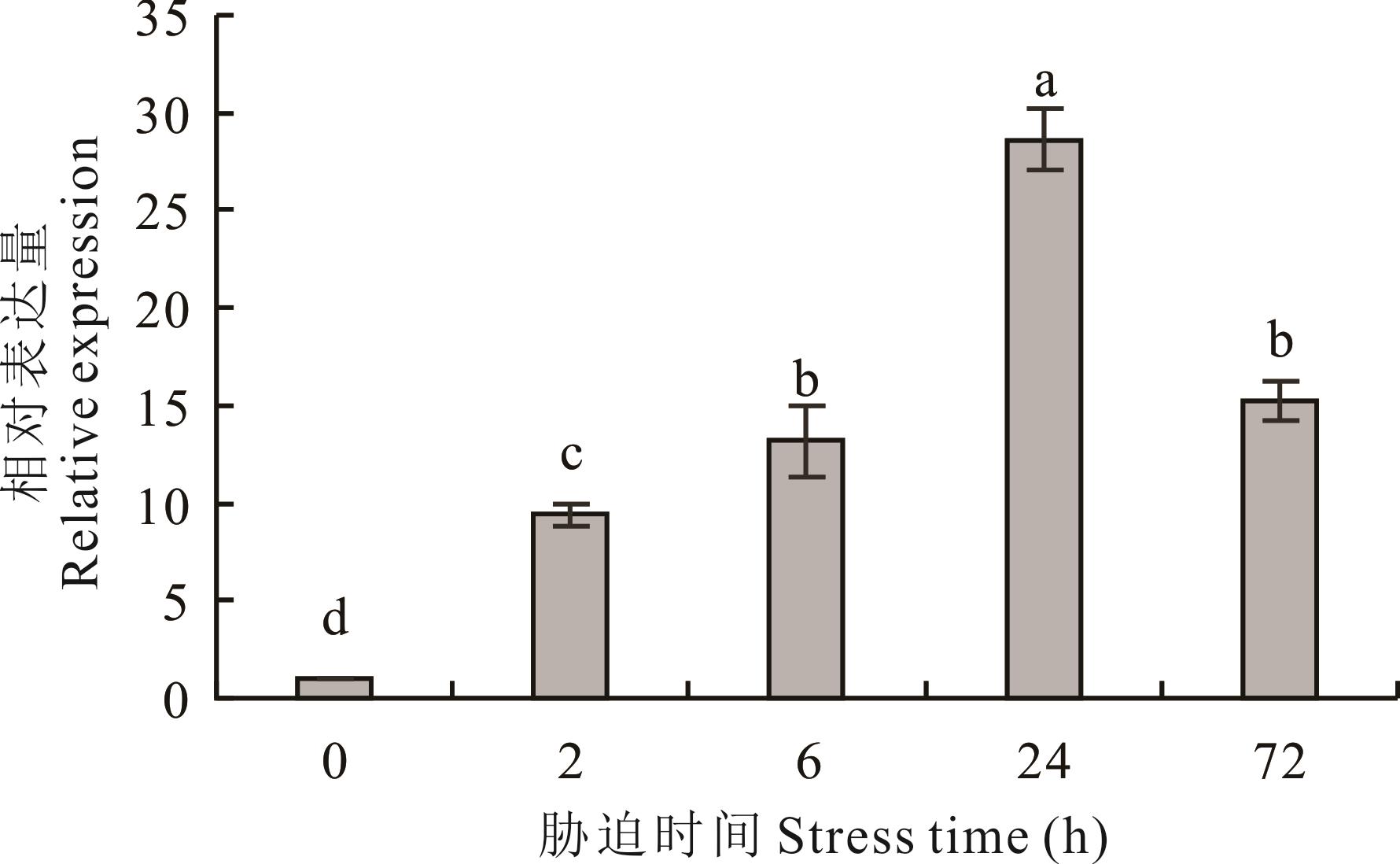
图 8 200 mmol·L-1 NaCl 胁迫不同时间下盐生草根系 HgAKR6C 的表达量不同小写字母表示差异显著(P<0.05),下同。Different lowercase letters indicate significant difference (P<0.05), the same below.
Fig.8 Expression of HgAKR6C in saltbush root systems under different times of 200 mmol·L-1 NaCl stress
| 1 | Daliakopoulos I N, Tsanis I K, Koutroulis A, et al. The threat of soil salinity: A European scale review. Science of the Total Environment, 2016, 573: 727-739. |
| 2 | Yang J S, Yao R J, Wang X P, et al. Research on salt-affected soils in China: History, status quo and prospect. Acta Pedologica Sinica, 2022, 59(1): 10-27. |
| 杨劲松, 姚荣江, 王相平, 等. 中国盐渍土研究: 历程、现状与展望. 土壤学报, 2022, 59(1): 10-27. | |
| 3 | Zhang H O. An analysis of the distribution and evolutionary characteristics of saline soils in China. Agriculture and Technology, 2022, 42(5): 104-107. |
| 张海欧. 浅析中国盐渍土分布及演变特征. 农业与技术, 2022, 42(5): 104-107. | |
| 4 | Ma H Y, Guo R, Li H A, et al. Study on salinity tolerance of tomatoes during seed germination under different salt stress conditions. Journal of Anhui Agricultural Sciences, 2008, 36(32): 13947-13948, 13956. |
| 马洪英, 郭锐, 李洪安, 等. 不同盐胁迫处理下番茄种子萌发期的耐盐性研究. 安徽农业科学, 2018, 36(32): 13947-13948, 13956. | |
| 5 | Pan J, Huang C H, Luo J, et al. Effects of salt stress on plant and the mechanism of arbuscular mycorrhizal fungi enhancing salt tolerance of plants. Advances in Earth Science, 2018, 33(4): 361-372. |
| 潘晶, 黄翠华, 罗君, 等. 盐胁迫对植物的影响及AMF提高植物耐盐性的机制. 地球科学进展, 2018, 33(4): 361-372. | |
| 6 | Munns R, Day D A, Fricke W, et al. Energy costs of salt tolerance in crop plants. New Phytologist, 2020, 225(3): 1072-1090. |
| 7 | Zelm E V, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annual Review of Plant Biology, 2020, 71: 403-433. |
| 8 | Shah K, Dubey R S. Effect of cadmium on RNA level as well as activity and molecular forms of ribonuclease in growing rice seedlings. Plant Physiology and Biochemistry, 1995, 33(5): 577-584. |
| 9 | Pan Y, Weng J, Cao Y, et al. Functional coupling between the Kv1.1 channel and an aldo-keto reductase Kvβ1. Journal of Biological Chemistry, 2008, 283(13): 8634-8642. |
| 10 | Auiyawong B, Narawongsanont R, Tantitadapitak C. Characterization of AKR4C15, a novel member of aldo-keto reductase, in comparison with other rice AKR(s). The Protein Journal, 2017, 36(4): 257-269. |
| 11 | Penning T M. The aldo-keto reductases (AKRs): Overview. Chemico-Biological Interactions, 2015, 234(5): 236-246. |
| 12 | Giuseppe P O D, Santos M L D, Sousa S M D, et al. A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases. Biochemical and Biophysical Research Communications, 2016, 474(4): 696-701. |
| 13 | Sengupta D, Naik D, Reddy A R. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. Journal of Plant Physiology, 2015, 179(1): 40-55. |
| 14 | Simpson P J, Tantitadapitak C, Reed A M, et al. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. Journal of Molecular Biology, 2009, 392(2): 465-480. |
| 15 | Narawongsanont R, Kabinpong S, Auiyawong B, et al. Cloning and characterization of AKR4C14, a rice aldo-keto reductase, from Thai Jasmine rice. The Protein Journal, 2012, 31(1): 35-42. |
| 16 | Editorial Committee of Flora Reipublicae Popularis Sinicae, Chinese Academy of Sciences. Flora reipublicae popularis sinicae. Beijing: Science Press, 1979. |
| 中国科学院中国植物志编辑委员会. 中国植物志. 北京: 科学出版社, 1979. | |
| 17 | Sun H Y, Zhang X M, Li L, et al. Estimation on aboveground biomass and the characteristics of population families of the halophilous herbaceous plants in three different areas of south Tarim Basin. Journal of Arid Land Resources and Environment, 2008(4): 193-197. |
| 孙红叶, 张希明, 李利, 等. 塔里木盆地南缘不同生境盐生草种群分布特征及地上生物量初步估测. 干旱区资源与环境, 2008(4): 193-197. | |
| 18 | Munns R. Genes and salt tolerance: Bringing them together. New Phytologist, 2005, 167(3): 645-663. |
| 19 | Wang J C. Study on the salt tolerance mechanisms of ion compartmentation in halophyte Halogeton glomeratus. Lanzhou: Gansu Agricultural University, 2017. |
| 汪军成. 盐生草盐分区隔化耐盐机制研究. 兰州: 甘肃农业大学, 2017. | |
| 20 | Yao L R. Study on the salt uptake mechanisms of roots in halophyte Halogeton glomeratus. Lanzhou: Gansu Agricultural University, 2018. |
| 姚立蓉. 盐生草根系对盐分吸收机理的研究. 兰州: 甘肃农业大学, 2018. | |
| 21 | Wang J C, Yao L R, Li B C, et al. Single-molecule long-read transcriptome dataset of halophyte Halogeton glomeratus. Frontiers in Genetics, 2017, 8: 197. |
| 22 | Xu K. The novel plant Na+/H+ antiporter gene evolved by DNA shuffling confers yeast and Arabidopsis improved salt tolerance. Shanghai: East China Normal University, 2010. |
| 徐凯. 通过体外DNA改组技术获得新的植物强耐盐Na+/H+逆向转运蛋白基因. 上海: 华东师范大学, 2010. | |
| 23 | Henning P M, Roalson E H, Mir W, et al. Annotation of the Turnera subulata (Passifloraceae) draft genome reveals the S-locus evolved after the divergence of turneroideae from Passifloroideae in a stepwise manner. Plants, 2023, 12(2): 286. |
| 24 | Rajewski A, Carter-House D, Stajich J, et al. Datura genome reveals duplications of psychoactive alkaloid biosynthetic genes and high mutation rate following tissue culture. BMC Genomics, 2021, 22(1): 1-19. |
| 25 | Tang H, Vasconcelos A C, Ma J, et al. In vivo expression pattern of a plant K+ channel β subunit. Plant Science, 1998, 134(2): 117-128. |
| 26 | Johnson A R, Yue Y, Carey S B, et al. Chromosome-level genome assembly of Euphorbia peplus, a model system for plant latex, reveals that relative lack of Ty3 transposons contributed to its small genome size. Genome Biology and Evolution, 2022, 15(3): evad018. |
| 27 | Blom N, Gammol S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. Journal of Molecular Biology, 1999, 294(5): 1351-1362. |
| 28 | Wang D, Hao Z, Zhao J, et al. Comparative physiological and transcriptomic analyses reveal salt tolerance mechanisms of Zygosaccharomyces rouxii. Process Biochemistry, 2019, 82: 59-67. |
| 29 | Wang D, Zhang M, Huang J, et al. Heat preadaptation improved the ability of Zygosaccharomyces rouxii to salt stress: A combined physiological and transcriptomic analysis. Applied Microbiology and Biotechnology, 2021, 10(5): 259-270. |
| 30 | Wong M M, Bhaskara G B, Wen T N, et al. Phosphoproteomics of Arabidopsis highly ABA-induced1 identifies AT-Hook-Like10 phosphorylation required for stress growth regulation. Proceedings of the National Academy of Sciences, 2019, 116(6): 2354-2363. |
| 31 | Chen Q, Xu X Y, Wang J C, et al. Identification of a WRKY gene family based on full-length transcriptome sequences and analysis of response patterns under salt stress in Halogeton glomeratus. Acta Prataculturae Sinica, 2022, 31(12): 146-157. |
| 陈倩, 徐晓芸, 汪军成, 等. 基于全长转录组的盐生草WRKY基因家族的鉴定及其盐胁迫响应模式分析. 草业学报, 2022, 31(12): 146-157. | |
| 32 | Wu P M, Cheng B, Leng Y, et al. Analysis of aldo-keto reductase genes in Vigna radiata and its response to cadmium stress. Journal of Lanzhou Jiaotong University, 2023, 42(1): 118-126. |
| 吴萍民, 程斌, 冷艳, 等. 绿豆醛酮还原酶基因及其响应镉胁迫的分析. 兰州交通大学学报, 2023, 42(1): 118-126. | |
| 33 | Mgobozi V, Afolayan A J, Otunola G A. Heavy metal uptake potential of Egeria densa (Plach) Casp. South African Journal of Botany, 2016, 103: 331-332. |
| 34 | Yu J, Sun H, Zhang J, et al. Analysis of aldo-keto reductase gene family and their responses to salt, drought, and abscisic acid stresses in Medicago truncatula. International Journal of Molecular Sciences, 2020, 21(3): 754. |
| 35 | Schachtman D P, Schroeder J, Lucas W J, et al. Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science, 1992, 258(5088): 1654-1658. |
| 36 | Tang H, Vasconcelos A C, Berkowitz G A. Evidence that plant K+ channel proteins have two different types of subunits. Plant Physiology, 1995, 109(1): 327-330. |
| 37 | Jan L Y, Jan Y N. Potassium channels and their evolving gates. Nature, 1994, 371(6493): 119-122. |
| 38 | Sussman M R. Shaking Arabidopsis thaliana. Science, 1992, 256(5057): 619. |
| 39 | Amtmann A, Fischer M, Marsh E L, et al. The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain. Plant Physiology, 2001, 126(3): 1061-1071. |
| 40 | Mäser P, Eckelman B, Vaidyanathan R, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Letters, 2002, 531(2): 157-161. |
| 41 | Fan L, Sun T J, Yang J, et al. Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene (GmNHX1) from soybean. Journal of Agricultural University of Hebei, 2015, 38(6): 7-12. |
| 范龙, 孙天杰, 杨郡, 等. 大豆GmNHX1基因克隆及其在酵母中的耐盐性分析. 河北农业大学学报, 2015, 38(6): 7-12. | |
| 42 | Guo H. The molecular basis of Atriplex canescens, a secretohalophyte with salt bladders, in response to NaCl. Lanzhou: Lanzhou University, 2020. |
| 郭欢. 盐囊泡类泌盐植物四翅滨藜响应NaCl的分子基础研究. 兰州: 兰州大学, 2020. | |
| 43 | Wang W Y. Functional characterization of HKT transporters in the succulent xerophyte Zygophyllum xanthoxylum. Lanzhou: Lanzhou University, 2019. |
| 王文颖. 多浆旱生植物霸王HKT转运蛋白的功能研究. 兰州: 兰州大学, 2019. | |
| 44 | Zhang H W, Xiao W, Yu W W, et al. Halophytic Hordeum brevisubulatum HbHAK1 facilitates potassium retention and contributes to salt tolerance. International Journal of Molecular Sciences, 2020, 21(15): 5292. |
| [1] | 王园, 王晶, 李淑霞. 紫花苜蓿MsBBX24基因的克隆及耐盐性分析[J]. 草业学报, 2023, 32(3): 107-117. |
| [2] | 刘福, 陈诚, 张凯旋, 周美亮, 张新全. 日本百脉根LjbHLH34基因克隆及耐旱功能鉴定[J]. 草业学报, 2023, 32(1): 178-191. |
| [3] | 陈倩, 徐晓芸, 汪军成, 姚立蓉, 司二静, 杨轲, 韦晓玲, 马小乐, 李葆春, 尚勋武, 孟亚雄, 王化俊. 基于全长转录组的盐生草WRKY基因家族的鉴定及其盐胁迫响应模式分析[J]. 草业学报, 2022, 31(12): 146-157. |
| [4] | 侯洁茹, 段晓玥, 李州, 彭燕. 白三叶TrSAMDC1克隆及表达分析[J]. 草业学报, 2020, 29(8): 170-178. |
| [5] | 罗维, 舒健虹, 刘晓霞, 王子苑, 牟琼, 王小利, 吴佳海. 高羊茅FaRVE8基因的克隆、亚细胞定位及表达分析[J]. 草业学报, 2020, 29(7): 60-69. |
| [6] | 杨婷, 张建平, 刘自刚, 齐燕妮, 李闻娟, 谢亚萍. 胡麻异质型ACCase亚基基因的克隆与表达分析[J]. 草业学报, 2020, 29(4): 111-120. |
| [7] | 何建军, 姚立蓉, 汪军成, 边秀秀, 司二静, 杨轲, 王化俊, 马小乐, 李葆春, 尚勋武, 孟亚雄. 干旱和盐胁迫对盐生植物盐生草种子萌发特性的影响[J]. 草业学报, 2020, 29(11): 129-140. |
| [8] | 夏曾润, 王文颖, 刘亚琪, 王锁民. 罗布麻K+通道编码基因AvAKT1的克隆与表达分析[J]. 草业学报, 2019, 28(8): 180-189. |
| [9] | 胡娜, 李葆春, 姚立蓉, 汪军成, 边秀秀, 侯静静, 司二静, 杨轲, 孟亚雄, 马小乐, 王化俊. 不同重金属胁迫对盐生草种子萌发特性的影响[J]. 草业学报, 2019, 28(6): 66-81. |
| [10] | 邹兰, 杨轲, 徐先良, 汪军成, 任盼荣, 姚立蓉, 孟亚雄, 李葆春, 马小乐, 王化俊. 盐生草HgNHX1基因启动子的克隆及功能验证[J]. 草业学报, 2017, 26(11): 57-68. |
| [11] | 杨毅, 陆姗姗, 刘萍, 田蕾. 苦豆子赖氨酸脱羧酶基因克隆与表达分析[J]. 草业学报, 2016, 25(8): 128-135. |
| [12] | 邵麟惠, 郑兴卫, 李聪. 蒺藜苜蓿E3泛素连接酶U-box基因克隆及表达分[J]. 草业学报, 2016, 25(7): 62-72. |
| [13] | 魏树强, 孙振元, 代小梅, 钱永强. 多年生黑麦草LpGCS基因克隆及其在烟草中的初步功能验证[J]. 草业学报, 2016, 25(4): 121-132. |
| [14] | T-DNA插入位点侧翼序列的克隆. 柱花草炭疽菌致病力丧失突变菌株1869的[J]. 草业学报, 2015, 24(8): 142-149. |
| [15] | 雒军,王引权,温随超,李静,张金林,夏琦. 当归苯丙氨酸解氨酶基因片段克隆和组织特异性表达分析[J]. 草业学报, 2014, 23(4): 130-137. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||