

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2026, Vol. 35 ›› Issue (1): 154-169.DOI: 10.11686/cyxb2025122
• 研究论文 • 上一篇
未丽1( ), 邓育轩1, 赵静2, 刘俊良2, 马克华2, 王锁民1(
), 邓育轩1, 赵静2, 刘俊良2, 马克华2, 王锁民1( )
)
收稿日期:2025-04-08
修回日期:2025-05-21
出版日期:2026-01-20
发布日期:2025-11-13
通讯作者:
王锁民
作者简介:E-mail: smwang@lzu.edu.cn基金资助:
Li WEI1( ), Yu-xuan DENG1, Jing ZHAO2, Jun-liang LIU2, Ke-hua MA2, Suo-min WANG1(
), Yu-xuan DENG1, Jing ZHAO2, Jun-liang LIU2, Ke-hua MA2, Suo-min WANG1( )
)
Received:2025-04-08
Revised:2025-05-21
Online:2026-01-20
Published:2025-11-13
Contact:
Suo-min WANG
摘要:
角质层蜡质在旱生植物霸王抵御不良生境中发挥着重要作用。β-酮脂酰辅酶A合成酶(KCS)是催化蜡质前体物质合成的关键酶。本研究克隆得到霸王ZxKCS6/CER6的cDNA全长1548 bp,具有2个决定底物选择性的高度保守跨膜区和3个酶催化位点,属于缩合酶超家族成员。与其他高等植物同源蛋白的氨基酸序列比对发现,ZxCER6属于KCS家族中CER6亚家族,具有第224位的半胱氨酸高度保守激活位点。系统进化树分析发现,ZxCER6与拟南芥AtCER6的亲缘关系最近。表达模式分析表明,ZxCER6主要在地上部组织中表达,尤其在叶表皮中表达丰度最高;且受50 mmol·L-1 NaCl处理的强烈诱导,其表达丰度在36 h时达到峰值。利用表皮特异性启动子驱动ZxCER6在拟南芥中表达发现,干旱处理后转基因拟南芥的地上部干鲜重、叶绿素含量、净光合速率和水分利用效率均显著高于野生型,而离体叶片失水率、叶绿素浸出率和叶片相对质膜透性均低于野生型;转基因植株地上部角质层蜡质含量,尤其是烷烃含量显著增加。表明ZxCER6的超表达可增加转基因植株表皮中烷烃的含量,减少水分散失,从而提高其抗旱性。本研究初步揭示了ZxCER6介导的蜡质积累在荒漠植物霸王抗旱性中的作用,为优良牧草及农作物抗旱性的遗传改良提供了优异的基因资源。
未丽, 邓育轩, 赵静, 刘俊良, 马克华, 王锁民. 旱生植物霸王ZxCER6的基因克隆及功能分析[J]. 草业学报, 2026, 35(1): 154-169.
Li WEI, Yu-xuan DENG, Jing ZHAO, Jun-liang LIU, Ke-hua MA, Suo-min WANG. Cloning and functional analysis of ZxCER6 from the xerophyte Zygophyllum xanthoxylum[J]. Acta Prataculturae Sinica, 2026, 35(1): 154-169.
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 目的Purpose |
|---|---|---|
| 3F-CER6 | ACGAGGCGGAGACTGTTATTT | 基因克隆 Gene cloning |
| 3FN-CER6 | ATCCCAATTCAAATGCTGTCG | |
| 5R-CER6 | CTTCATACACGCAACGATAGG | |
| 5RN-CER6 | TCATCAGCTCCTTTGTGGGTC | |
| WF-CER6 | AACAGTCCACTGCCTTCAACAGTAC | |
| WR-CER6 | GCCATCCACCAATCAACCTTAT | |
| P | CTAATACGACTCACTATAGGGC | |
| NP | AAGCAGTGGTATCAACGCAGAGT | |
| QF-ZxCER6 | CGTTGCGTGTATGAAGAAGAGG | qRT-PCR |
| QR-ZxCER6 | TATGGCTTGATTTTCGGGTTG | |
| QF-ZxACTIN | TTTTCCAGCCATCCCTTGTT | |
| QR-ZxACTIN | TGCAGTGATCTCCTTGCTCATAC | |
| VF AtCER6(HindⅢ) | TGTTGGCCCAAGCTTCTTCGATATCGGTTGTTGACGAT | 植物表达载体构建 Construction of plant expression vector |
| VR AtCER6 | CAAGATTTGAGGCATCGTCGGAGAGTTTTAATGTATAAT | |
| VF ZxCER6 | ATGCCTCAAATCTTGCCCGATTTCT | |
| VR ZxCER6(SacⅠ) | GGGAAATTCGAGCTCCTACAGCTTGACAACTTCAGGAAT |
表1 所用引物信息
Table 1 Primer information used in the experiment
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) | 目的Purpose |
|---|---|---|
| 3F-CER6 | ACGAGGCGGAGACTGTTATTT | 基因克隆 Gene cloning |
| 3FN-CER6 | ATCCCAATTCAAATGCTGTCG | |
| 5R-CER6 | CTTCATACACGCAACGATAGG | |
| 5RN-CER6 | TCATCAGCTCCTTTGTGGGTC | |
| WF-CER6 | AACAGTCCACTGCCTTCAACAGTAC | |
| WR-CER6 | GCCATCCACCAATCAACCTTAT | |
| P | CTAATACGACTCACTATAGGGC | |
| NP | AAGCAGTGGTATCAACGCAGAGT | |
| QF-ZxCER6 | CGTTGCGTGTATGAAGAAGAGG | qRT-PCR |
| QR-ZxCER6 | TATGGCTTGATTTTCGGGTTG | |
| QF-ZxACTIN | TTTTCCAGCCATCCCTTGTT | |
| QR-ZxACTIN | TGCAGTGATCTCCTTGCTCATAC | |
| VF AtCER6(HindⅢ) | TGTTGGCCCAAGCTTCTTCGATATCGGTTGTTGACGAT | 植物表达载体构建 Construction of plant expression vector |
| VR AtCER6 | CAAGATTTGAGGCATCGTCGGAGAGTTTTAATGTATAAT | |
| VF ZxCER6 | ATGCCTCAAATCTTGCCCGATTTCT | |
| VR ZxCER6(SacⅠ) | GGGAAATTCGAGCTCCTACAGCTTGACAACTTCAGGAAT |
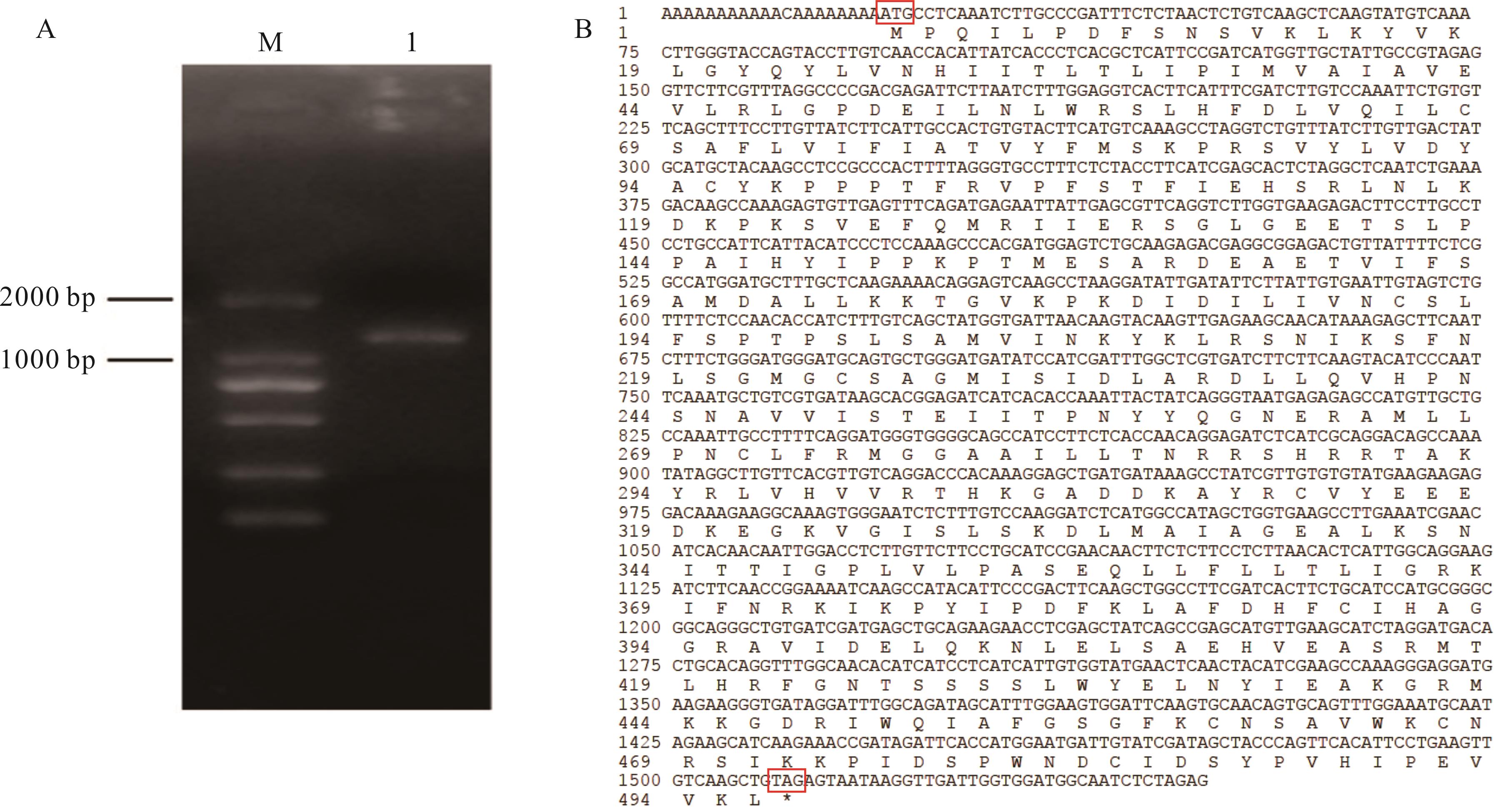
图1 ZxCER6的克隆与cDNA全长及对应蛋白序列A: ZxCER6基因的PCR扩增产物PCR amplification product of ZxCER6 gene; M: DL 2000 DNA marker; 下同The same below. 1: cDNA扩增产物cDNA amplified product. B: ZxCER6的cDNA全长序列Full cDNA sequence of ZxCER6. 红色方框处为起始密码子(ATG)和终止密码子(TAG)。The start codon (ATG) and the stop codon (TAG) were marked in red boxes.
Fig.1 Cloning full-length cDNA and corresponding protein sequence of ZxCER6
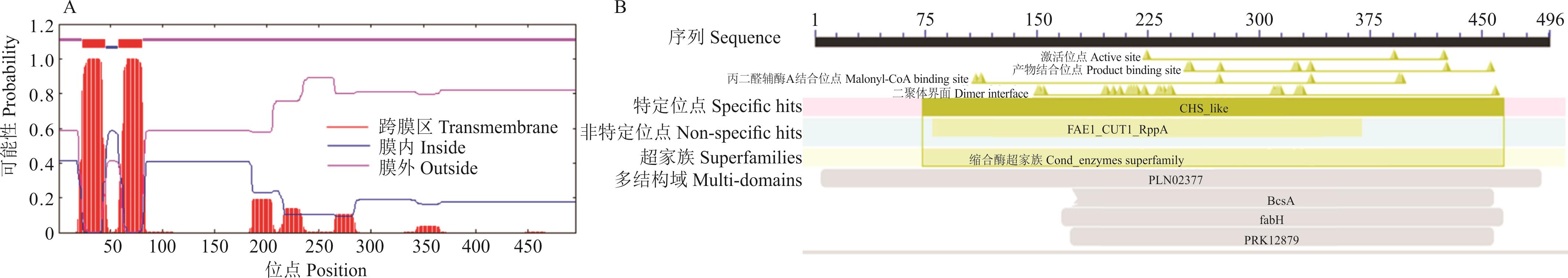
图2 ZxCER6蛋白结构特征分析A: ZxCER6蛋白的跨膜域ZxCER6 protein transmembrane domain; B: ZxCER6蛋白保守结构域ZxCER6 protein conserved domain.
Fig.2 Structural characterization analysis of ZxCER6 protein

图3 ZxCER6与其他高等植物CER6的氨基酸序列多重比对CmCER6: 甜瓜Cucumis melo CER6 (XP_008446598.1); CsCER6: 黄瓜Cucumis sativus CER6 (XP_004135090.1); EgCER6: 巨桉Eucalyptus grandis CER6 (XP_010066726.1); GmCER6: 大豆Glycine max CER6 (XP_003555901.1); PmCER6: 梅Prunus mume CER6 (XP_008243224.1); PtCER6: 毛果杨Populus trichocarpa CER6 (XP_002311457.1); TcCER6: 可可Theobroma cacao CER6 (XP_007044356.2); ZxCER6:霸王Z. xanthoxylum CER6. 下同The same below. 黄色方框表示CER6的2个跨膜区(TM1、TM2),红色星号表示高度保守激活位点Cys224。Transmembrane domain (TM1, TM2) was indicated with yellow box. A highly conserved catalytically residue was indicated with red asterisk.
Fig.3 Amino acid sequence alignment of ZxCER6 with other CER6 from higher plants
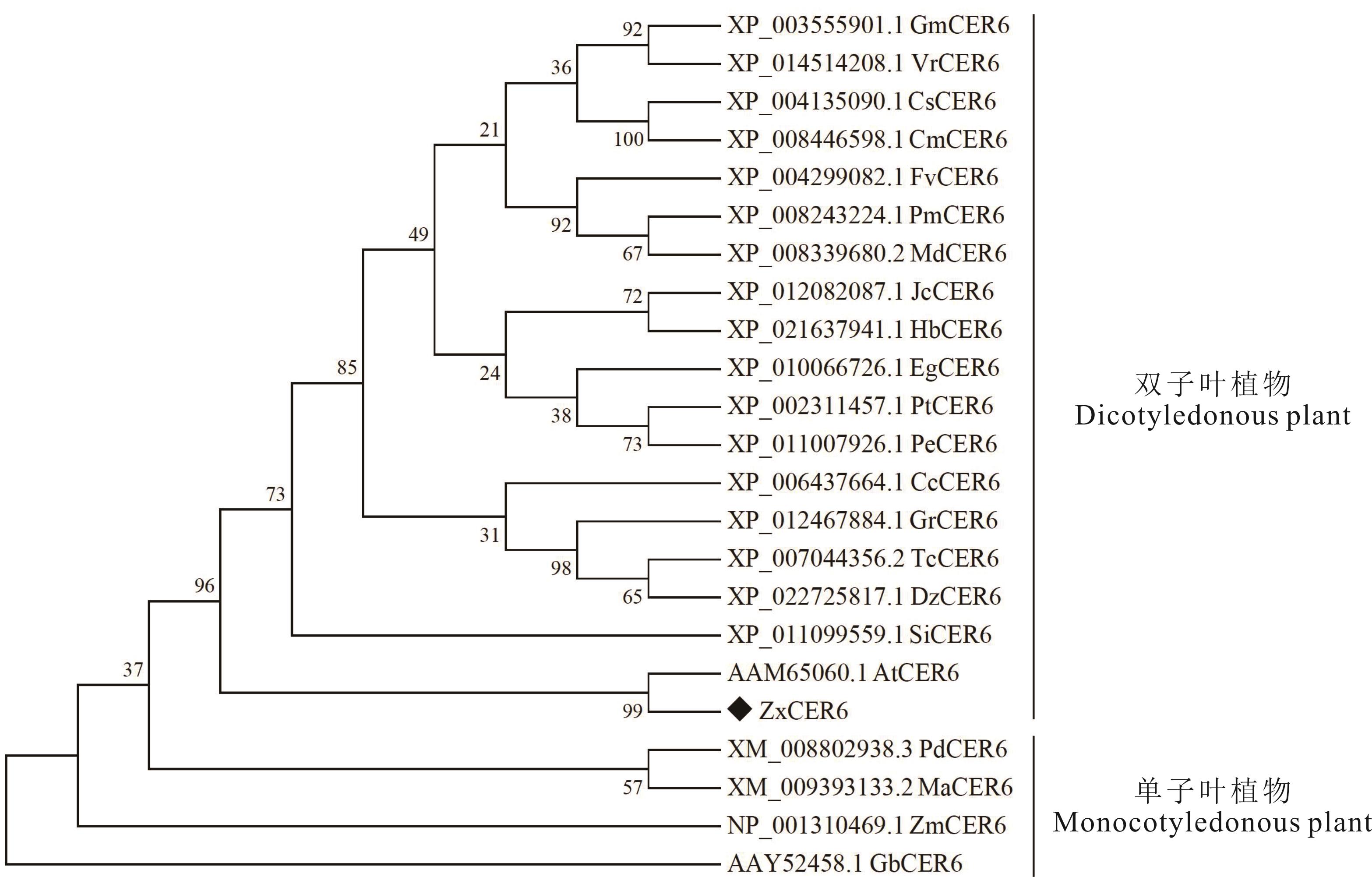
图4 不同植物CER6系统进化树分析VrCER6: 三裂叶绿豆Vigna radiata var. radiata CER6; FvCER6: 林地草莓Fragaria vesca subsp. vesca CER6; MdCER6: 苹果Malus domestica CER6; JcCER6: 麻疯树Jatropha curcasx CER6; HbCER6: 橡胶树Hevea brasiliensis CER6; PeCER6: 胡杨Populus euphratica CER6; CcCER6: 克莱门柚Citrus clementina CER6; GrCER6: 雷蒙德氏棉G. raimondii CER6; DzCER6: 榴莲Durio zibethinus CER6; SiCER6: 芝麻Sesamum indicum CER6; AtCER6: 拟南芥A. thaliana CER6; PdCER6: 海枣P. dactylifera CER6; MaCER6: 小果野芭蕉M. acuminate CER6; ZmCER6: 玉米Zea mays CER6; GbCER6: 银杏Ginkgo biloba CER6.
Fig.4 Phylogenetic tree analysis of CER6 in different plants
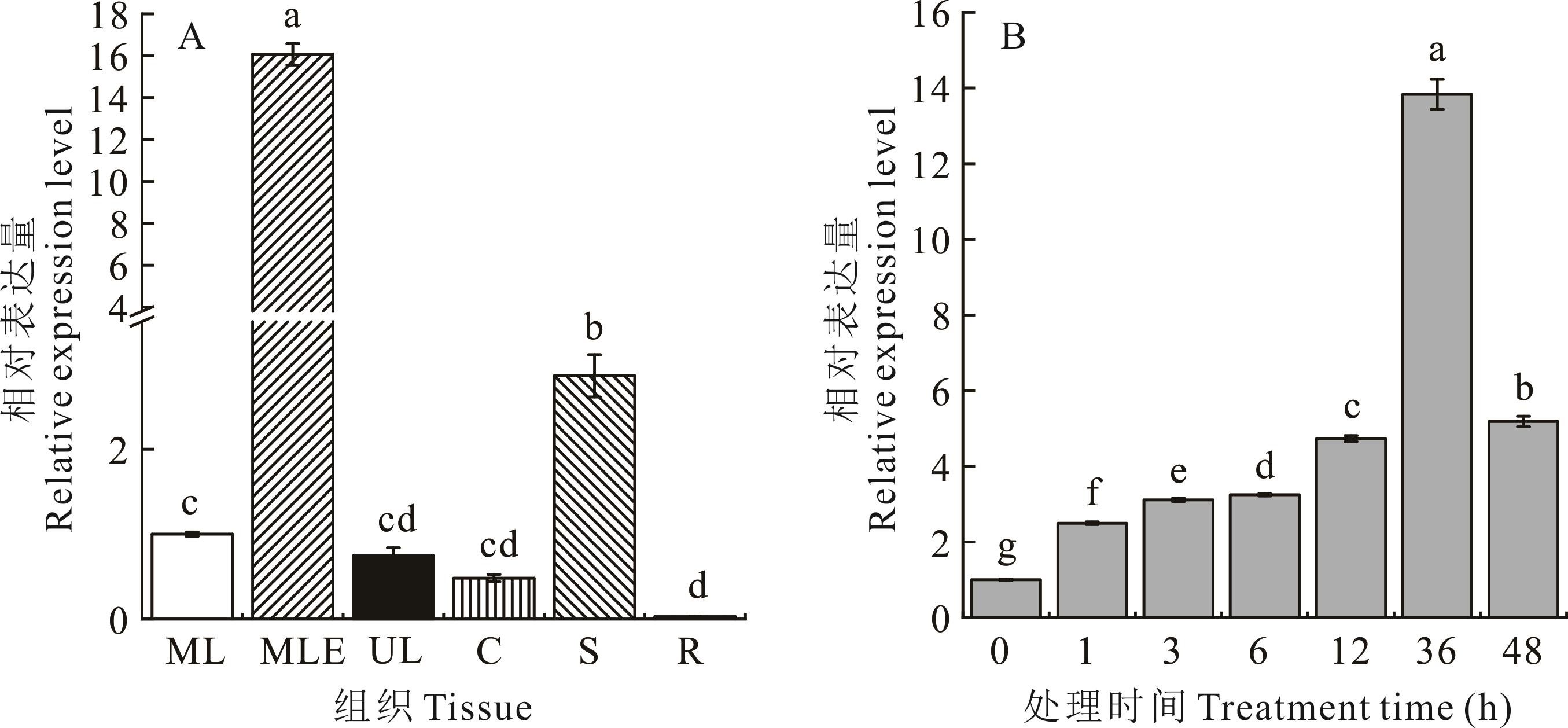
图5 ZxCER6的表达模式分析A: 组织特异性表达模式Tissue specific expression pattern; ML: 中层叶Middle leaves; MLE: 中层叶表皮Epidermis of middle leaves; UL: 上层叶Upper leaves; C: 子叶Cotyledons; S: 茎Stems; R: 根Roots. B: 50 mmol·L-1 NaCl处理下表达模式Expression patterns under 50 mmol·L-1 NaCl. 不同小写字母表示不同部位或不同处理时间下差异显著(P<0.05)。Different lowercase letters indicate significant difference among different tissues or different treatments time (P<0.05).
Fig.5 Expression pattern analysis of ZxCER6
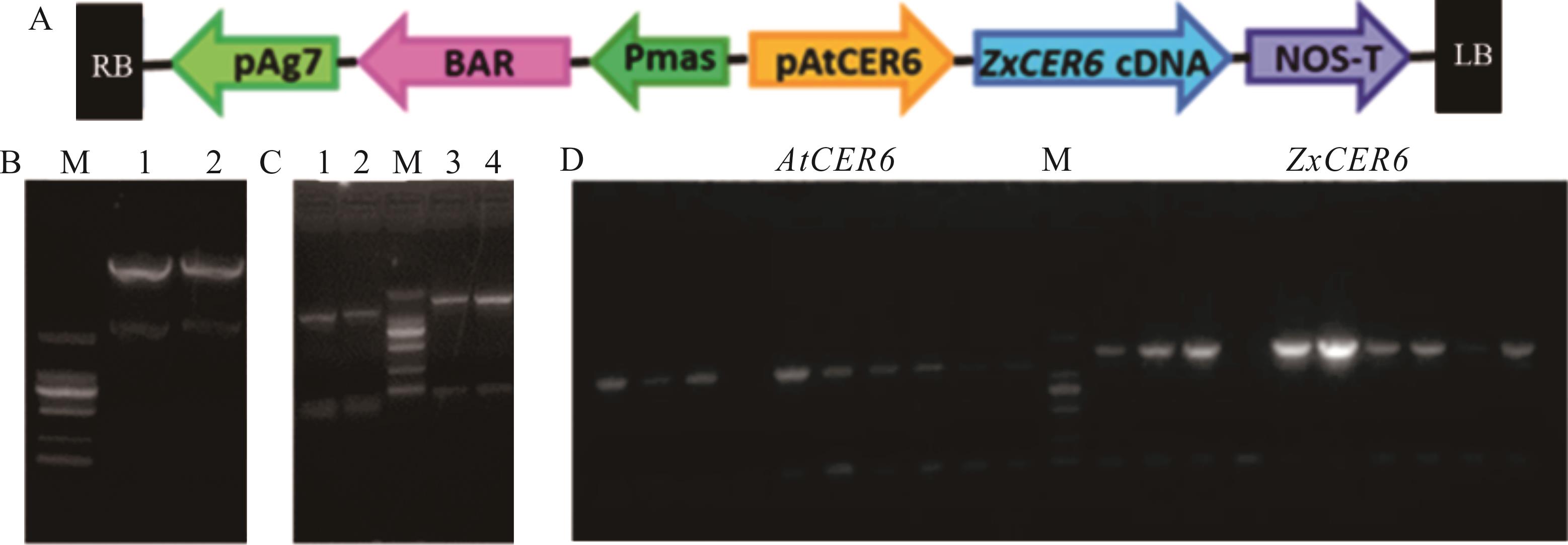
图6 植物表达载体的构建A: 植物表达载体pBIB-BASTA-pAtCER6::ZxCER6构建示意图Schematic diagram of plant expression vector pBIB-BASTA-pAtCER6::ZxCER6 construction. B: pBIB-BASTA-35S-GWR-FLAG基础载体Hind Ⅲ和Sac Ⅰ双酶切电泳图谱pBIB-BASTA-35S-GWR-FLAGHind Ⅲand Sac Ⅰ double enzyme electrophoresis; 1, 2: 双酶切电泳条带The electrophoretic bands of double enzyme. C: AtCER6和ZxCER6分别加入Hind Ⅲ和Sac Ⅰ酶切位点电泳图谱AtCER6 and ZxCER6 join Hind Ⅲ and Sac Ⅰ enzyme electrophoresis; 1, 2: AtCER6电泳条带AtCER6 electrophoretic bands; 3, 4: ZxCER6电泳条带ZxCER6 electrophoretic bands. D: AtCER6和ZxCER6菌液PCR电泳图谱PCR electrophoresis of AtCER6 and ZxCER6 strains.
Fig.6 Construction of plant expression vector
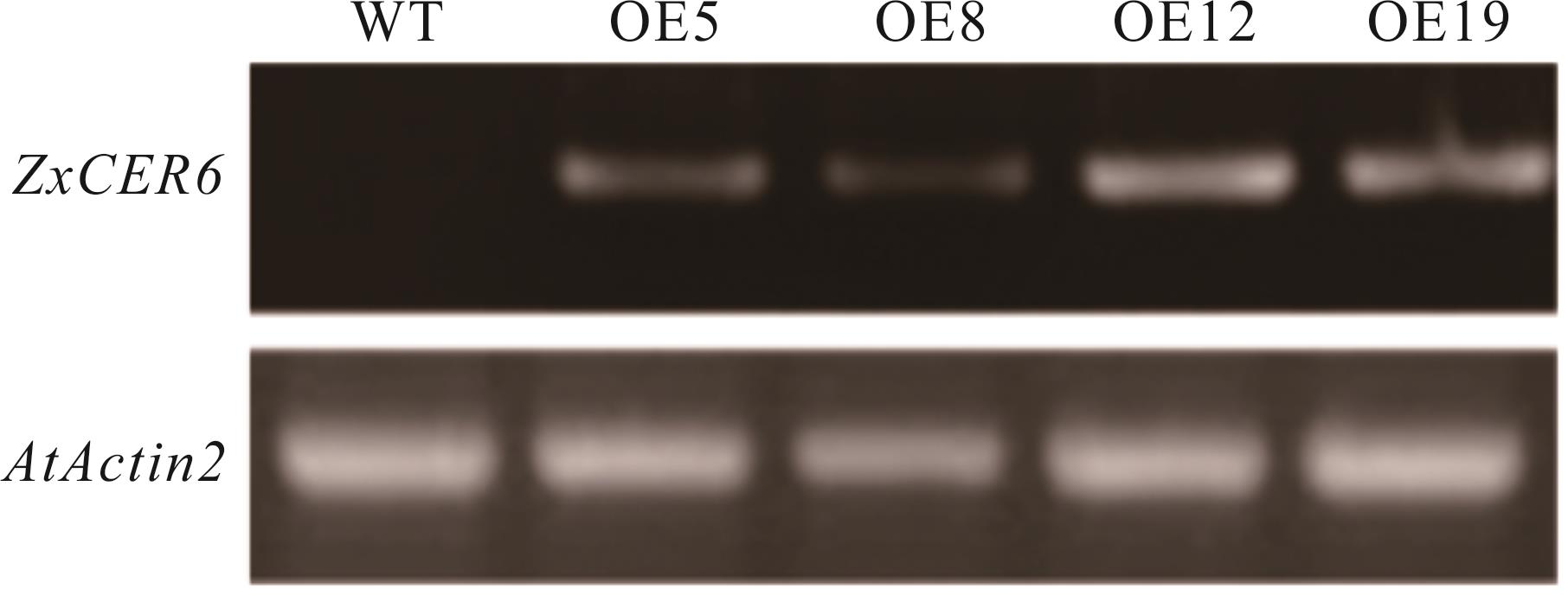
图7 转基因拟南芥中ZxCER6 的RT-PCR检测WT: 野生型拟南芥植株Wild type Arabidopsis plants; OE5, OE8, OE12, OE19: 转基因拟南芥株系Transgenic Arabidopsis lines; AtActin2: 内参基因Internal gene. 下同The same below.
Fig.7 RT-PCR detection of ZxCER6 in transgenic Arabidopsis
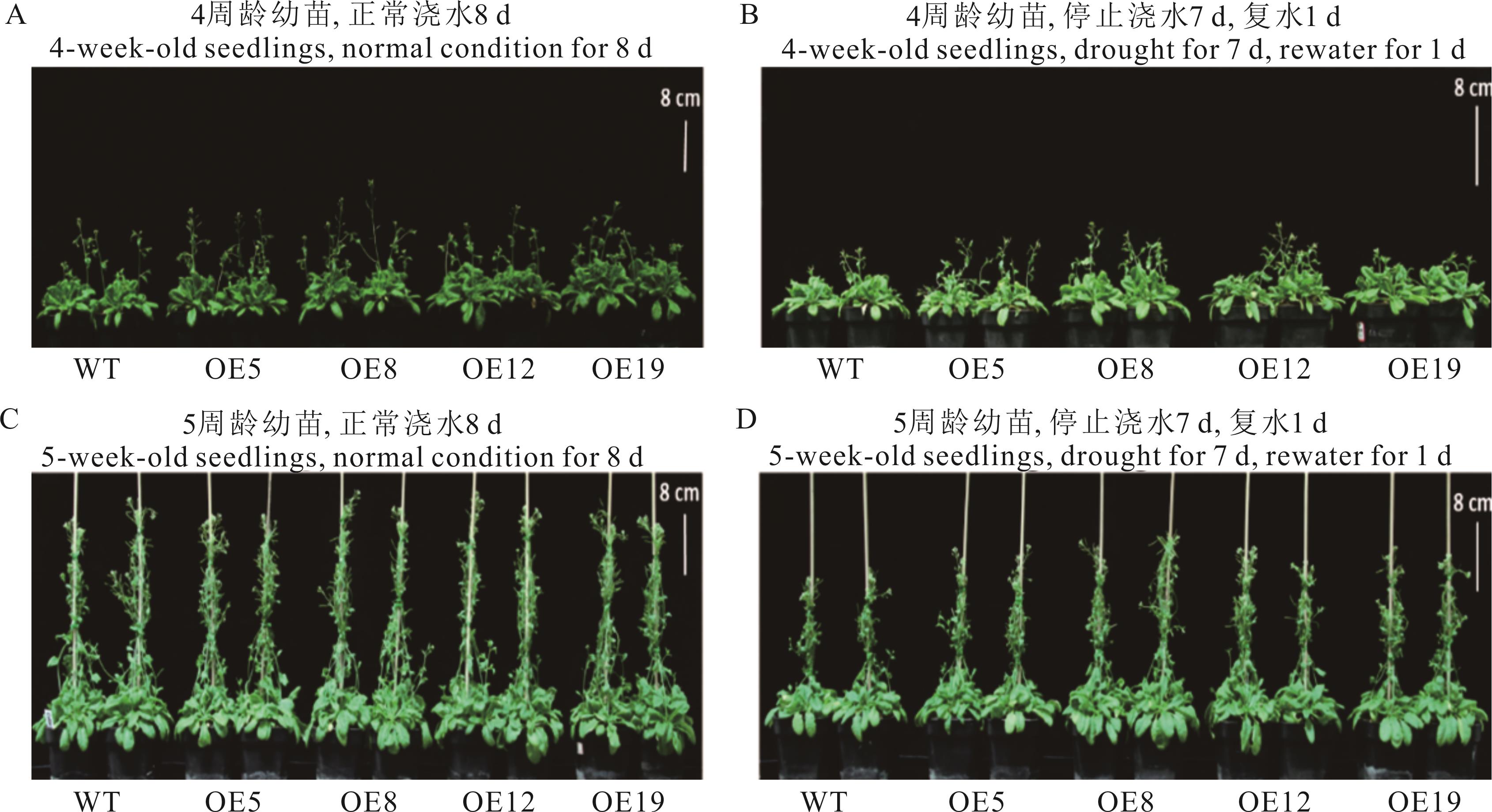
图8 不同干旱周期下野生型和ZxCER6转基因拟南芥的生长状态图A和B分别为第1个干旱周期对照及处理;图C和D分别为第2个干旱周期对照及处理。A: The first drought period control; B: The first drought period; C: The second drought period control; D: The second drought period.
Fig.8 The growth state of wild type and ZxCER6 transgenic Arabidopsis under different drought periods

图9 不同干旱周期下野生型和ZxCER6转基因拟南芥植株株高A: 第1个干旱周期The first drought period; B: 第2个干旱周期The second drought period. 不同小写字母表示野生型和不同转基因株系在不同处理间差异显著(P<0.05)。Different lowercase letters indicate that WT and different transgenic lines have significant differences among different treatments (P<0.05). 下同The same below.
Fig.9 The plant height of wild type and ZxCER6 transgenic Arabidopsis under different drought periods

图11 第2个干旱周期下野生型和ZxCER6转基因拟南芥叶片叶绿素含量和光合参数分析
Fig.11 Analysis of chlorophyll content and photosynthesis index between wide type and ZxCER6 transgenic Arabidopsis in the second drought period
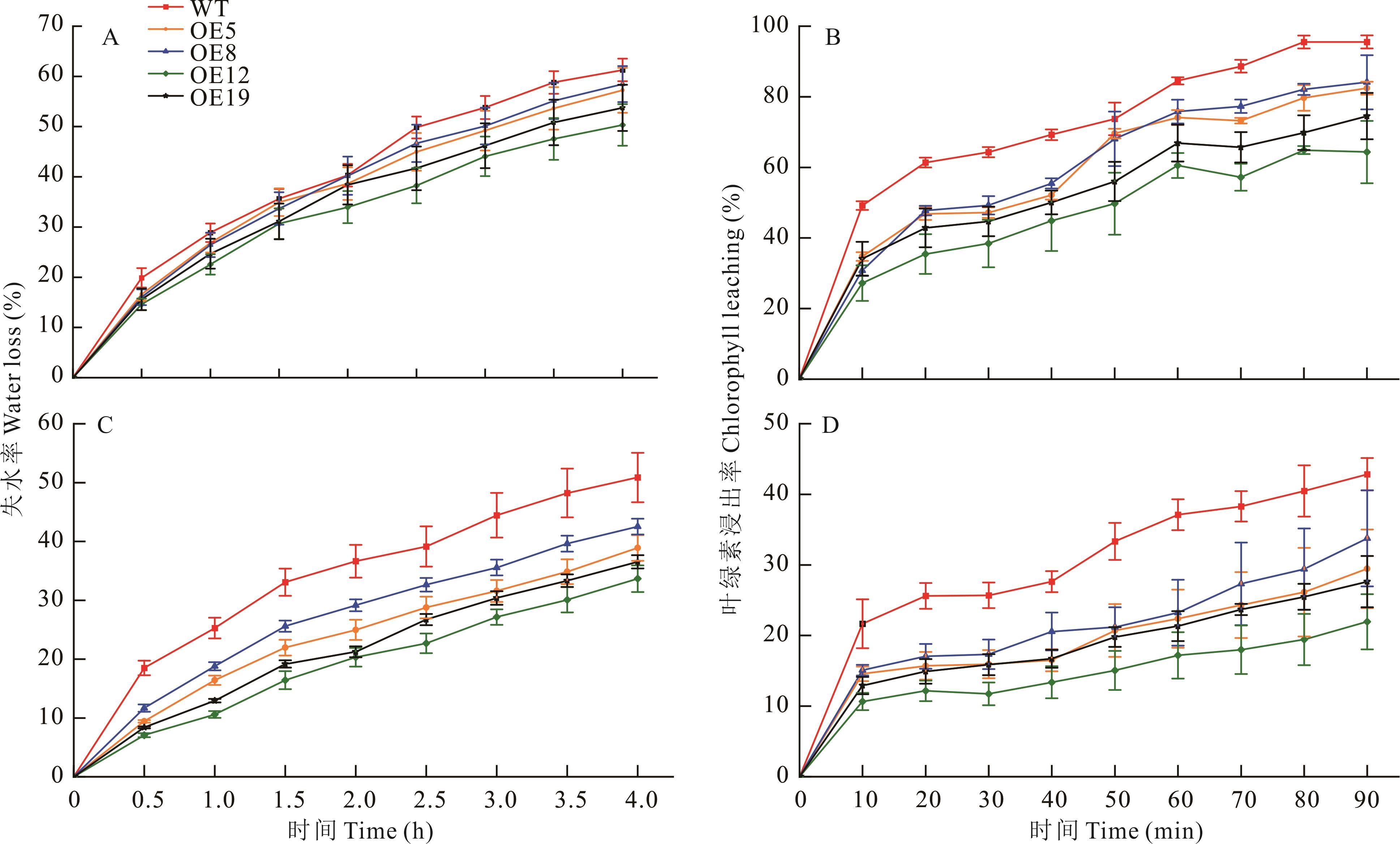
图12 野生型和ZxCER6转基因拟南芥叶片的角质层透性正常条件(A、B)和第2个干旱周期下(C、D)野生型和ZxCER6转基因拟南芥叶片失水率(A、C)及叶绿素浸出率(B、D)。Water loss rate (A, C) and chlorophyll leaching rate (B, D) of wild type and ZxCER6 transgenic Arabidopsis leaves under control (A, B) and the second drought period (C, D). 数值为平均值±标准误(n=5)。Data are mean±standard error (n=5).
Fig.12 Cuticle permeability of wide type and ZxCER6 transgenic Arabidopsis leaves
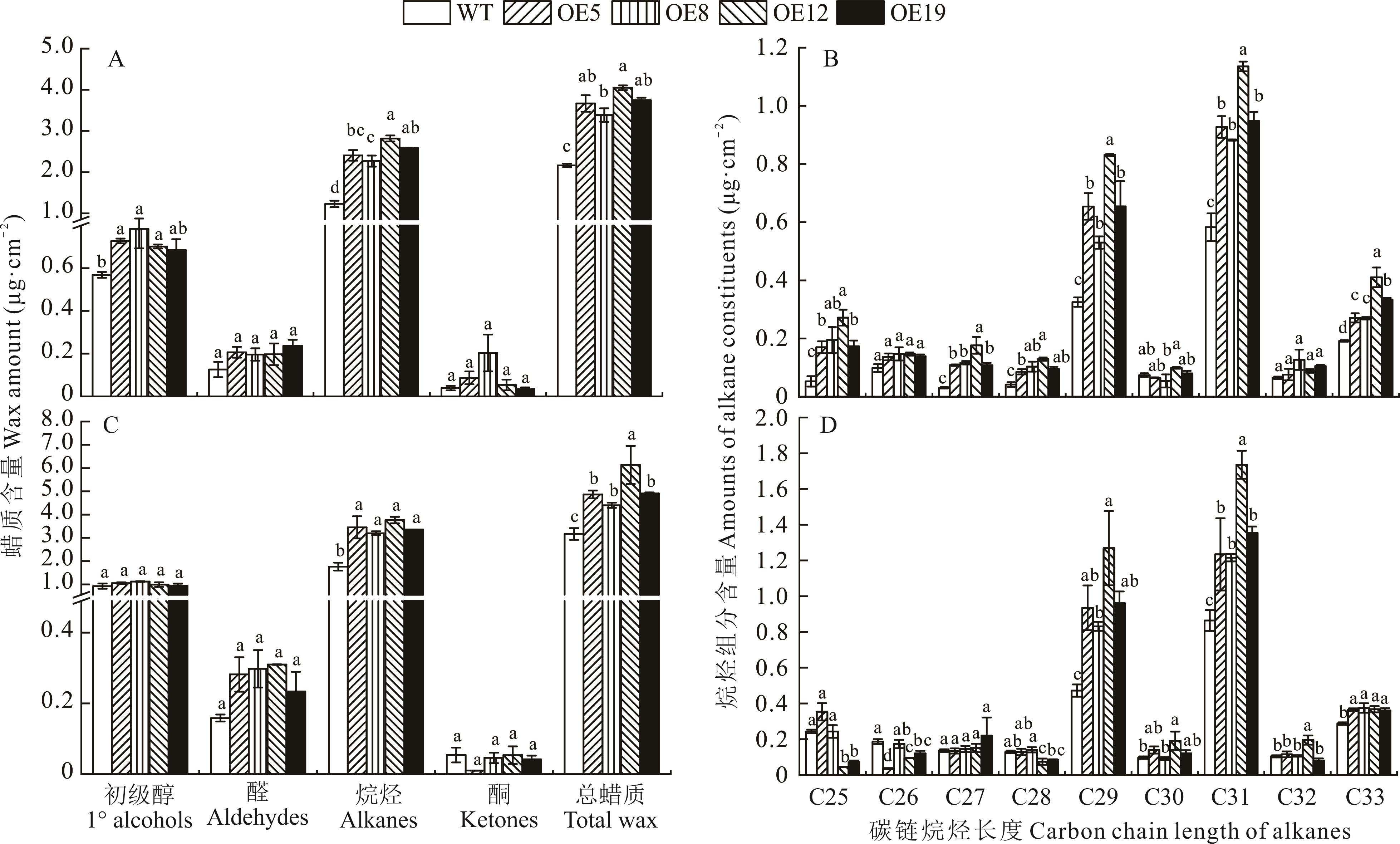
图15 野生型和ZxCER6转基因拟南芥莲座叶中角质层蜡质含量及不同碳链长度烷烃组分含量正常条件(A、B)和第2个干旱周期下(C、D)野生型和ZxCER6转基因拟南芥莲座叶角质层蜡质含量(A、C)及不同碳链长度烷烃组分含量(B、D)。Wax amount (A, C) and amounts of different chain alkane constituents (B, D) on rosette leaves of wild type and ZxCER6 transgenic Arabidopsis under control (A, B) and the second drought period (C, D). 不同小写字母表示各株系之间差异显著(P<0.05)。下同。Different lowercase letters indicate significant difference among the lines (P<0.05). The same below.
Fig.15 Wax amount and amounts of different chain alkane constituents on rosette leaves of wide type and ZxCER6 transgenic Arabidopsis
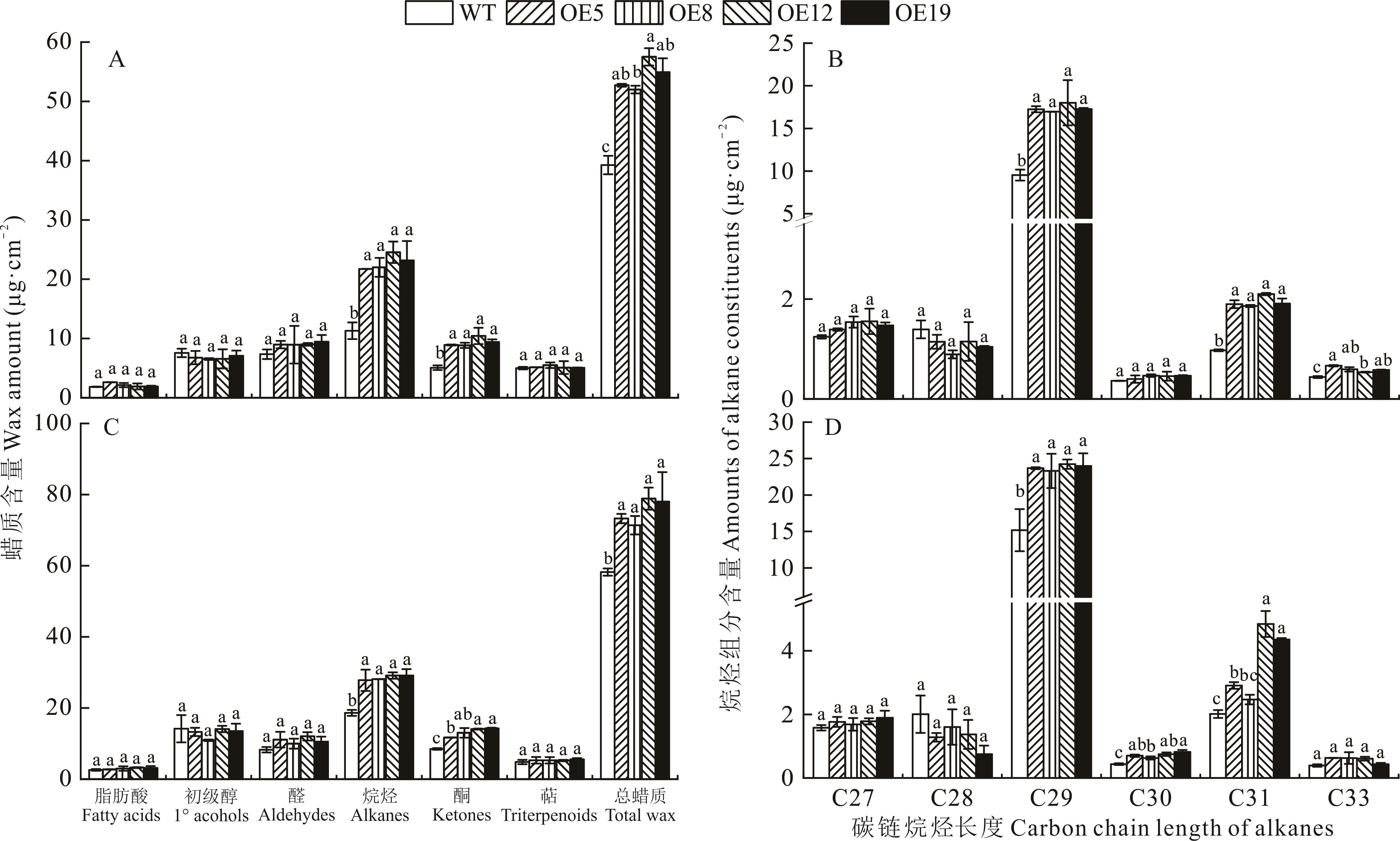
图16 野生型和ZxCER6转基因拟南芥花序茎角质层蜡质含量及不同碳链长度烷烃组分含量正常条件(A、B)和第2个干旱周期下(C、D)野生型和ZxCER6转基因拟南芥花序茎角质层蜡质含量(A、C)及不同碳链长度烷烃组分含量(B、D)。Wax amount (A, C) and amounts of different chain alkane constituents (B, D) on inflorescence stems of wild type and ZxCER6 transgenic Arabidopsis under control (A, B) and the second drought period (C, D).
Fig.16 Wax amount and amounts of different chain alkane constituents on inflorescence stems of wide type and ZxCER6 transgenic Arabidopsis
| [1] | Rossak M, Smith M, Kunst L. Expression of the FAE1 gene and FAE1 promoter activity in development seeds of Arabidopsis thaliana. Plant Molecular Biology, 2001, 46(6): 717-725. |
| [2] | Costaglioli P, Joubès J, Garcia C, et al. Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2005, 1734(3): 247-258. |
| [3] | Joubès J, Raffaele S, Bourdenx B, et al. The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modeling and expression profiling. Plant Molecular Biology, 2008, 67(5): 547-566. |
| [4] | Todd J, Post-Beittenmiller D, Jaworski J G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. The Plant Journal, 1999, 17(2): 119-130. |
| [5] | Lee S B, Jung S J, Go Y S, et al. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. The Plant Journal, 2009, 60(3): 462-475. |
| [6] | Pruitt R E, Vielle-Calzada J P, Ploense S E, et al. FIDDLEHEAD, a gene required to suppress epidermal cell interaction in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proceedings of the National Academy of the Sciences of the United States of America, 2000, 97(3): 1311-1316. |
| [7] | Gray J E, Holroyd G H, van der Lee F M, et al. The HIC signalling pathway links CO2 perception to stomatal development. Nature, 2000, 408(6813): 713-716. |
| [8] | Fiebig A, Mayfield J A, Chau S, et al. Alterations in CER6, a gene identical to CUT1, differentially affect long chain lipid content on the surface of pollen and stems. Plant Cell, 2000, 12(10): 2001-2008. |
| [9] | Jenks M A, Tuttle H A, Eigenbrode S D, et al. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiology, 1995, 108(1): 369-377. |
| [10] | Hooker T S, Millar A A, Kunst L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiology, 2002, 129(4): 1568-1580. |
| [11] | Leide J, Hildebrandt U, Reussing K, et al. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: Effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiology, 2007, 144(3): 1667-1679. |
| [12] | Richardson A, Boscari A, Schreiber L, et al. Cloning and expression analysis of candidate genes involved in wax deposition along the growing barley (Hordeum vulgare) leaf. Planta, 2007, 226(6): 1459-1473. |
| [13] | Qin Y M, Pujol F M, Hu C Y, et al. Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid enlongation. Journal of Experimental Botany, 2007, 58(3): 473-481. |
| [14] | Li H T. Cloning and characterization of the ThCER6 gene from Thellungiella halophila. Jinan: Shandong Normal University, 2004. |
| 李宏韬. 小盐芥(Thellungiella halophila)ThCER6基因的克隆与功能研究. 济南: 山东师范大学, 2004. | |
| [15] | Wu Y H, Cao Y L, Luo J L, et al. Cloning and functional characterization of KCS6 genes from Brassica napus. Chinese Journal of Oil Crop Sciences, 2012, 34(6): 567-574. |
| 武玉花, 曹应龙, 罗军玲, 等. 甘蓝型油菜中KCS6基因的克隆和功能分析. 中国油料作物学报, 2012, 34(6): 567-574. | |
| [16] | Krauss P, Markstädter C, Riederer M. Attenuation of UV radiation by plant cuticles from woody species. Plant Cell Environment, 1997, 20(8): 1079-1085. |
| [17] | Sieber P, Schorderet M, Ryser U, et al. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. The Plant Cell, 2000, 12(5): 721-737. |
| [18] | Karaba A. Improvement of water use efficiency in rice and tomato using Arabidopsis wax biosynthetic genes and transcription factors. Wageningen: Wageningen University & Research, 2007. |
| [19] | Sun Y Y. Improvement of drought and salt tolerance using Arabidopsis BOUNTIFUL and HARDY gene and epicuticular wax synthesis genes in tomato. Beijing: Chinese Academy of Agricultural Sciences, 2012. |
| 孙玉燕. 利用拟南芥BOUNTIFUL和HARDY基因以及表皮蜡质合成基因提高番茄耐旱和耐盐性. 北京: 中国农业科学院, 2012. | |
| [20] | Zhang J Y, Broeckling C D, Blancaflor E B, et al. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). The Plant Journal, 2005, 42(5): 689-707. |
| [21] | Jiang Q, Zhang J Y, Guo X, et al. Physiological characterization of transgenic alfalfa (Medicago sativa) plant for improved drought tolerance. International Journal of Plant Sciences, 2009, 170(8): 969-978. |
| [22] | Wang H H, Hao J J, Chen X J, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Molecular Biology, 2007, 65(6): 799-815. |
| [23] | Islam M A, Du H, Ning J, et al. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Molecular Biology, 2009, 70(4): 443-456. |
| [24] | Wang Y H. Rice DRF2, a transcriptional activator, is involved in leaf wax synthesis. Beijing: Chinese Academy of Agricultural Sciences, 2010. |
| 王友华. 水稻ERF转录激活子DRF2调控叶表蜡质合成. 北京: 中国农业科学院, 2010. | |
| [25] | Zhang Y L, Zhang C L, Wang G L, et al. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biology, 2019, 19(1): 362. |
| [26] | Wang Y M, Jin S R, Xu Y, et al. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop Journal, 2020, 8(1): 26-37. |
| [27] | Ma Q, Yue L J, Zhang J L, et al. Sodium chloride improves photosynthesis and water status in the succulent xerophyte Zygophyllum xanthoxylum. Tree Physiology, 2012, 32(1): 4-13. |
| [28] | Zhao C X, Huang Z C. A preliminary study of xeromorphism of some important xerophytes growing in Tungeli Desert. Acta Botanica Sinica, 1981, 23(4): 278-283. |
| 赵翠仙, 黄子琛. 腾格里沙漠主要旱生植物旱性结构的初步研究. 植物学报, 1981, 23(4): 278-283. | |
| [29] | Ma Y L, Wang L, Liu Y X, et al. Updates on stress tolerance of main accessory structures and their synergetic interaction in desert plants. Plant Physiology Journal, 2015, 51(11): 1821-1836. |
| 马亚丽, 王璐, 刘艳霞, 等. 荒漠植物几种主要附属结构的抗逆功能及其协同调控的研究进展. 植物生理学报, 2015, 51(11): 1821-1836. | |
| [30] | Li H J, Bai W P, Liu L B, et al. Massive increases in C31 alkane on Zygophyllum xanthoxylum leaves contribute to its excellent abiotic stress tolerance. Annals of Botany, 2023, 131(4): 723-736. |
| [31] | Tian Y. Functional verification of ZxABCG11 from Zygophyllum xanthoxylum and genetic transformation to alfalfa (Medicago sativa L.). Lanzhou: Lanzhou University, 2017. |
| 田野. 霸王ZxABCG11的功能验证及其对紫花苜蓿(Medicago sativa L.)的遗传转化. 兰州: 兰州大学, 2017. | |
| [32] | Lolle S J, Berlyn G P, Engstrom E M, et al. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Development Biology, 1997, 189(2): 311-321. |
| [33] | Kosma D K, Bourdenx B, Bernard A, et al. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology, 2009, 151(4): 1918-1929. |
| [34] | Millar A A, Clemens S, Zachgo S, et al. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell, 1999, 11(5): 825-838. |
| [35] | Vogg G, Fischer S, Leide J, et al. Tomato fruit cuticular waxes and the effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. Journal of Experimental Botany, 2004, 55(401): 1401-1410. |
| [36] | Wang X C, Guan Y Y, Zhang D, et al. A beta-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiology, 2017, 173(2): 944-955. |
| [37] | Lassner M W, Lardizabal K, Metz J G. A jojoba beta-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutant in transgenic plants. Plant Cell, 1996, 8(2): 281-292. |
| [38] | Ghanevati M, Jaworski J G. Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase, FAE1 KCS. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2001, 1530(1): 77-85. |
| [39] | Busta L, Budke J M, Jetter R. Identification of β-hydroxy fatty acid esters and primary, secondary-alkanediol esters in cuticular waxes of the moss Funaria hygrometrica. Phytochemistry, 2016, 121(1): 38-49. |
| [40] | Suh M C, Samuels A L, Jetter R, et al. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology, 2005, 139(4): 1649-1665. |
| [41] | Aharoni A, Dixit S, Jetter R, et al. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. The Plant Cell, 2004, 16(9): 2463-2480. |
| [42] | Bourdenx B, Bernard A, Domergue F, et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiology, 2011, 156(1): 29-45. |
| [43] | Zhang J Y, Broeckling C D, Sumner L W, et al. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Molecular Biology, 2007, 64(3): 265-278. |
| [44] | Lee S B, Kim H, Kim R J, et al. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Reports, 2014, 33(9): 1535-1546. |
| [45] | Lokesh U, Venkatesh B, Kiranmai K, et al. Overexpression of β-ketoacyl-CoA synthase1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) cultivar K-6 by increased leaf epicuticular wax accumulation. Frontiers in Plant Science, 2019, 9(11): 1869. |
| [1] | 蒋学乾, 杨青川, 康俊梅. 紫花苜蓿在干旱胁迫下的产量损失与抗旱性遗传研究进展[J]. 草业学报, 2025, 34(7): 219-234. |
| [2] | 赵媛媛, 蒲小剑, 徐成体, 王伟, 傅云洁. 蒺藜苜蓿MtBMI1基因克隆及抗旱性分析[J]. 草业学报, 2025, 34(6): 139-153. |
| [3] | 陈彩锦, 包明芳, 王文虎, 尚继红, 曾燕霞, 沙晓弟, 朱新忠, 王学敏, 刘文辉. 紫花苜蓿抗旱育种研究现状及展望[J]. 草业学报, 2025, 34(3): 204-223. |
| [4] | 汪欣瑶, 彭亚萍, 姚立蓉, 汪军成, 司二静, 张宏, 杨轲, 马小乐, 孟亚雄, 王化俊, 李葆春. 盐生草HgS5基因的克隆与抗旱性鉴定[J]. 草业学报, 2025, 34(2): 184-195. |
| [5] | 王少鹏, 刘佳, 洪军, 林积圳, 张义, 史昆, 王赞. 紫花苜蓿MsPPR1基因的克隆及抗旱功能分析[J]. 草业学报, 2023, 32(7): 49-60. |
| [6] | 王文娟, 师尚礼, 何龙, 武蓓, 刘旵旵. 干旱胁迫下多胺在植物体内的积累及其作用[J]. 草业学报, 2023, 32(6): 186-202. |
| [7] | 姚露花, 綦才, 杨建峰, 郭彦军. 种子引发对甜高粱角质层蜡质及其抗性的影响[J]. 草业学报, 2022, 31(7): 185-196. |
| [8] | 畅志鹏, 孙莹莹, 李佳阳, 龚春梅. 柠条CkCAD基因的克隆转化及其抗旱功能分析[J]. 草业学报, 2021, 30(3): 68-80. |
| [9] | 蔺豆豆, 赵桂琴, 琚泽亮, 宫文龙. 15份燕麦材料苗期抗旱性综合评价[J]. 草业学报, 2021, 30(11): 108-121. |
| [10] | 何海锋, 闫承宏, 吴娜, 刘吉利, 贾瑜琀. 不同施氮水平对柳枝稷光合特性及抗旱性的影响[J]. 草业学报, 2021, 30(1): 107-115. |
| [11] | 张雪婷, 王新永, 杨文雄, 柳娜, 杨长刚. 河西绿洲灌区节水抗旱型玉米品种的评价方法探讨[J]. 草业学报, 2020, 29(2): 134-148. |
| [12] | 杨柳慧, 尹航, 黄沁梅, 张彦妮, 何淼, 周蕴薇. 细叶百合LpWRKY20基因对非生物胁迫的响应及抗旱性分析[J]. 草业学报, 2020, 29(1): 193-202. |
| [13] | 姜红岩, 滕珂, 檀鹏辉, 尹淑霞. 日本结缕草ZjZFN1基因对拟南芥的转化及其耐旱性分析[J]. 草业学报, 2019, 28(4): 129-138. |
| [14] | 郭星, 谢飞, 闫倩倩, 曹秀文, 杨帆. 黄腐酸对白龙江干旱河谷5种苗木抗旱性的影响[J]. 草业学报, 2018, 27(8): 86-94. |
| [15] | 刘婷婷, 陈道钳, 王仕稳, 殷俐娜, 邓西平. 不同品种高粱幼苗在干旱复水过程中的生理生态响应[J]. 草业学报, 2018, 27(6): 100-110. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||