

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (7): 111-121.DOI: 10.11686/cyxb2021215
• 研究论文 • 上一篇
收稿日期:2021-05-24
修回日期:2021-08-26
出版日期:2022-07-20
发布日期:2022-06-01
通讯作者:
刘文献
作者简介:E-mail: liuwx@lzu.edu.cn基金资助:
Jiao-yang TIAN( ), Qiu-xia WANG, Shu-wen ZHENG, Wen-xian LIU(
), Qiu-xia WANG, Shu-wen ZHENG, Wen-xian LIU( )
)
Received:2021-05-24
Revised:2021-08-26
Online:2022-07-20
Published:2022-06-01
Contact:
Wen-xian LIU
摘要:
CPP (cysteine-rich polycomb-like protein)基因家族是一类小转录因子,广泛存在于除酵母与原核生物外的各种生物体中,在植物生长发育及响应胁迫过程中具有重要作用。截至目前,CPP基因家族已在多个物种中被鉴定和研究,但在豆科模式植物蒺藜苜蓿中还未见报道。本研究采用生物信息学的方法,在蒺藜苜蓿中共鉴定出9个CPP基因。通过与3个物种CPP基因的蛋白序列构建系统发育树,将CPP基因分为3类,MtCPP成员与大豆的亲缘关系更接近。保守结构域分析表明MtCPP转录因子都具有1~2个CXC保守结构域。染色体定位分析得出,9个CPP基因分布在6条染色体上,并鉴定出2对旁系同源基因,均来源于片段重复事件。通过基因芯片技术检测MtCPP基因的表达模式结果显示,MtCPP基因的表达具有一定的时空特异性。MtCPP基因启动子中含有大量与激素以及胁迫相关的顺式作用元件,并且MtCPP2、MtCPP5、MtCPP6和MtCPP7基因具有响应干旱的表达特征,MtCPP2和MtCPP8基因具有响应盐胁迫的功能。该研究可为后期深入解析MtCPP基因家族功能和筛选参与苜蓿抗逆的MtCPP基因奠定基础。
田骄阳, 王秋霞, 郑淑文, 刘文献. 全基因组水平蒺藜苜蓿CPP基因家族的鉴定及表达模式分析[J]. 草业学报, 2022, 31(7): 111-121.
Jiao-yang TIAN, Qiu-xia WANG, Shu-wen ZHENG, Wen-xian LIU. Genome-wide identification and expression profile analysis of the CPP gene family in Medicago truncatula[J]. Acta Prataculturae Sinica, 2022, 31(7): 111-121.
序 号 No. | 基因名称 Gene name | Phytozome号 Phytozome ID | 编码序列 Coding sequence (bp) | 蛋白质 Protein (AA) | 染色体定位 Chromosomal location | 分子量 Molecular weight (Da) | 等电点 Isoelectric point | 亲水指数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MtCPP1 | Medtr1g012020.1 | 2604 | 867 | chr1:2312974..2319368 (-) | 93893.24 | 5.62 | -0.670 | 细胞核 Nucleus |
| 2 | MtCPP2 | Medtr1g086980.1 | 2496 | 831 | chr1:38931314..38941671 (+) | 93501.18 | 7.97 | -0.775 | 细胞核 Nucleus |
| 3 | MtCPP3 | Medtr1g103180.1 | 1101 | 366 | chr1:46677103..46682000 (-) | 39979.06 | 4.60 | -0.674 | 细胞核 Nucleus |
| 4 | MtCPP4 | Medtr1g103230.1 | 1875 | 624 | chr1:46702340..46705795 (-) | 70071.61 | 6.85 | -0.802 | 细胞核 Nucleus |
| 5 | MtCPP5 | Medtr2g049305.1 | 924 | 307 | chr2:21725271..21728509 (-) | 34610.89 | 8.50 | -0.169 | 叶绿体Chloroplast |
| 6 | MtCPP6 | Medtr3g110122.1 | 1791 | 596 | chr3:51048745..51054541 (-) | 65811.15 | 8.93 | -0.493 | 细胞核 Nucleus |
| 7 | MtCPP7 | Medtr5g006530.1 | 2337 | 778 | chr5:955306..960319 (-) | 85345.06 | 5.55 | -0.693 | 细胞核 Nucleus |
| 8 | MtCPP8 | Medtr6g087590.1 | 1824 | 607 | chr6:32942677..32948999 (+) | 65195.40 | 6.79 | -0.550 | 细胞核 Nucleus |
| 9 | MtCPP9 | Medtr8g103320.1 | 1437 | 478 | chr8:43471929..43476739 (+) | 51756.42 | 8.12 | -0.648 | 细胞核 Nucleus |
表1 MtCPP家族基本信息
Table 1 The basic information of MtCPP gene family in M. truncatula
序 号 No. | 基因名称 Gene name | Phytozome号 Phytozome ID | 编码序列 Coding sequence (bp) | 蛋白质 Protein (AA) | 染色体定位 Chromosomal location | 分子量 Molecular weight (Da) | 等电点 Isoelectric point | 亲水指数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MtCPP1 | Medtr1g012020.1 | 2604 | 867 | chr1:2312974..2319368 (-) | 93893.24 | 5.62 | -0.670 | 细胞核 Nucleus |
| 2 | MtCPP2 | Medtr1g086980.1 | 2496 | 831 | chr1:38931314..38941671 (+) | 93501.18 | 7.97 | -0.775 | 细胞核 Nucleus |
| 3 | MtCPP3 | Medtr1g103180.1 | 1101 | 366 | chr1:46677103..46682000 (-) | 39979.06 | 4.60 | -0.674 | 细胞核 Nucleus |
| 4 | MtCPP4 | Medtr1g103230.1 | 1875 | 624 | chr1:46702340..46705795 (-) | 70071.61 | 6.85 | -0.802 | 细胞核 Nucleus |
| 5 | MtCPP5 | Medtr2g049305.1 | 924 | 307 | chr2:21725271..21728509 (-) | 34610.89 | 8.50 | -0.169 | 叶绿体Chloroplast |
| 6 | MtCPP6 | Medtr3g110122.1 | 1791 | 596 | chr3:51048745..51054541 (-) | 65811.15 | 8.93 | -0.493 | 细胞核 Nucleus |
| 7 | MtCPP7 | Medtr5g006530.1 | 2337 | 778 | chr5:955306..960319 (-) | 85345.06 | 5.55 | -0.693 | 细胞核 Nucleus |
| 8 | MtCPP8 | Medtr6g087590.1 | 1824 | 607 | chr6:32942677..32948999 (+) | 65195.40 | 6.79 | -0.550 | 细胞核 Nucleus |
| 9 | MtCPP9 | Medtr8g103320.1 | 1437 | 478 | chr8:43471929..43476739 (+) | 51756.42 | 8.12 | -0.648 | 细胞核 Nucleus |
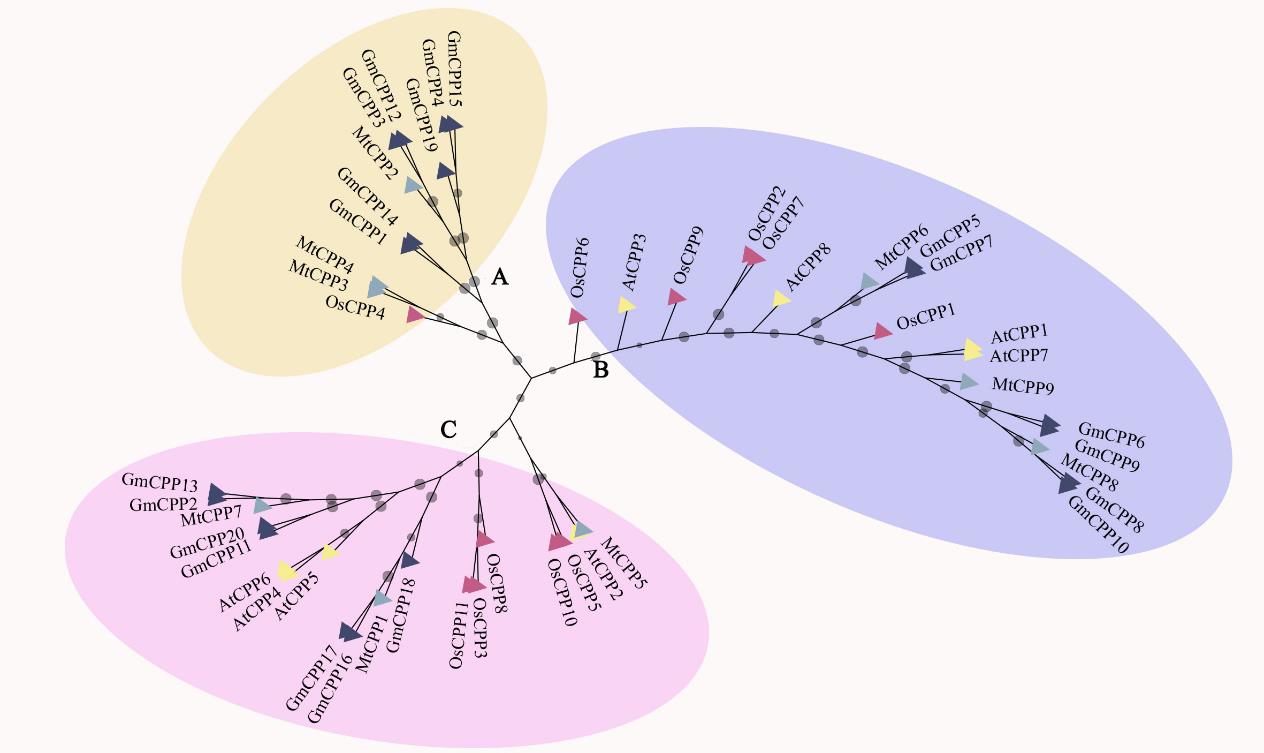
图1 MtCPP基因家族进化分析MtCPP为蒺藜苜蓿CPP蛋白,GmCPP为大豆CPP蛋白,OsCPP为水稻CPP蛋白,AtCPP为拟南芥CPP蛋白。MtCPP is the CPP protein of M. truncatula, GmCPPis the CPPprotein of soybean, OsCPP is the CPP protein of rice, AtCPP is the CPP protein of Arabidopsis.
Fig. 1 The phylogenetic analysis of MtCPP gene family
同源基因对位点 Homologous gene pairs | 非同义替换率 Ka | 同义替换率 Ks | Ka/Ks |
|---|---|---|---|
| MtCPP6/MtCPP9 | 0.48 | 2.87 | 0.17 |
| MtCPP8/MtCPP9 | 0.14 | 0.72 | 0.20 |
表2 MtCPP基因进化压力分析
Table 2 Evolutionary stress analysis of MtCPP genes
同源基因对位点 Homologous gene pairs | 非同义替换率 Ka | 同义替换率 Ks | Ka/Ks |
|---|---|---|---|
| MtCPP6/MtCPP9 | 0.48 | 2.87 | 0.17 |
| MtCPP8/MtCPP9 | 0.14 | 0.72 | 0.20 |
| 1 | Zhang L Q, Li H, Zhang F Y, et al. Research progress in Medicago transcription factors. Chinese Journal of Grassland, 2014, 36(2): 108-116. |
| 张立全, 李慧, 张凤英, 等. 苜蓿属植物转录因子研究进展. 中国草地学报, 2014, 36(2): 108-116. | |
| 2 | Wang B, Chen M D, Lin L, et al. Signal pathways and related transcription factors of drought stress in plants. Acta Botanica Boreali-Occidentalia Sinica, 2020, 40(10): 1792-1806. |
| 王彬, 陈敏氡, 林亮, 等. 植物干旱胁迫的信号通路及相关转录因子研究进展. 西北植物学报, 2020, 40(10): 1792-1806. | |
| 3 | Andersen S U, Algreen-Petersen R G, Hoedl M, et al. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. Journal of Experimental Botany, 2007, 58(13): 3657-3670. |
| 4 | Lu T, Dou Y, Zhang C. Fuzzy clustering of CPP family in plants with evolution and interaction analyses. BMC Bioinformatics, 2013, 14(13): 1-8. |
| 5 | Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence‐specific manner. Febs Journal, 2010, 276(19): 5703-5716. |
| 6 | Brzeska K, Brzeski J, Chandler S, et al. Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(12): 5516-5521. |
| 7 | Yang Z F, Gu S L, Wang X F, et al. Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. Journal of Molecular Evolution, 2008, 67(3): 266-277. |
| 8 | Zhang L, Zhao H K, Wang Y M, et al. Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genetics and Molecular Research, 2015, 14(1): 1260-1268. |
| 9 | Song X Y, Zhang Y Y, Wu F C, et al. Genome-wide analysis of the maize (Zea mays L.) CPP-like gene family and expression profiling under abiotic stress. Genetics and Molecular Research, 2016, 15(3): 1-11. |
| 10 | Zhou Y, Hu L F, Ye S F, et al. Genome-wide identification and characterization of cysteine-rich polycomb-like protein (CPP) family genes in cucumber (Cucumis sativus) and their roles in stress responses. Biologia, 2018, 73(4): 425-435. |
| 11 | Yang R X, Wang P J, Chen Z Z, et al. Genome-wide identification and analysis of CPP transcription factor family in tea plants. Acta Botanica Boreali-Occidentalia Sinica, 2019, 39(6): 1024-1032. |
| 杨如兴, 王鹏杰, 陈芝芝, 等. 茶树CPP转录因子家族的全基因组鉴定及分析. 西北植物学报, 2019, 39(6): 1024-1032. | |
| 12 | Liu Z C, Running M P, Meyerowitz E M. TSO1 functions in cell division during Arabidopsis flower development. Development, 1997, 124(3): 665-672. |
| 13 | Sijacic P, Wang W, Liu Z. Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PloS Genetics, 2011, 7(11): e1002352. |
| 14 | Cvitanich C, Pallisgaard N, Nielsen K A, et al. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proceedings of the National Academy of Sciences, 2000, 97(14): 8163-8168. |
| 15 | Wei Z W, Gai J Y. Model legume: Medicago truncatula. Acta Prataculturae Sinica, 2008, 17(1): 114-120. |
| 魏臻武, 盖钧镒. 豆科模式植物——蒺藜苜蓿. 草业学报, 2008, 17(1): 114-120. | |
| 16 | Wang Q X, Wei N, Jin X Y, et al. Molecular characterization of the COPT/Ctr-type copper transporter family under heavy metal stress in alfalfa. International Journal of Biological Macromolecules, 2021, 181: 644-652. |
| 17 | Jia X T, Liu W X, Xie W G, et al. Genome-wide analysis of the LBD transcription factor family in Medicago truncatula. Acta Botanica Boreali-Occidentalia Sinica, 2014, 34(11): 2176-2187. |
| 贾喜涛, 刘文献, 谢文刚, 等. 蒺藜苜蓿LBD转录因子基因家族全基因组分析. 西北植物学报, 2014, 34(11): 2176-2187. | |
| 18 | Min X Y, Wu H L, Zhang Z S, et al. Genome-wide identification and characterization of the aquaporin gene family in Medicago truncatula. Journal of Plant Biochemistry and Biotechnology, 2019, 28(3): 320-335. |
| 19 | Zhang Z S, Wei X Y, Liu W X, et al. Genome-wide identification and expression analysis of the fatty acid desaturase genes in Medicago truncatula. Biochemical and Biophysical Research Communications, 2018, 499(3): 361-367. |
| 20 | Finn R D, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Research, 2004, 36(Database issue): D281-D288. |
| 21 | Eddy S R. Accelerated profile HMM searches. PloS Computational Biology, 2011, 7(10): e1002195. |
| 22 | Panu A, Manohar J, Konstantin A, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 2012, 40(W1): W597-W603. |
| 23 | Chou K C, Shen H B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One, 2017, 5(6): e11335. |
| 24 | Newman L, Duffus A, Lee C. Using the free program MEGA to build phylogenetic trees from molecular data. The American Biology Teacher, 2016, 78(7): 608-612. |
| 25 | Chen C, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 26 | Wang Y, Tang H, Debarry J D, et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 2012, 40(7): e49. |
| 27 | Bailey T L, Mikael B, Buske F A, et al. Meme suite: Tools for motif discovery and searching. Nucleic Acids Research, 2009, 37(Web server issue): W202-W208. |
| 28 | Hurst L D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends in Genetics, 2002, 18(9): 486. |
| 29 | Wang K. Bioinformatic analysis of the CPP transcription factors family in Arabidopsis and rice. Biotechnology Bulletin, 2010(2): 76-84. |
| 王凯. 拟南芥和水稻CPP转录因子家族的生物信息学分析. 生物技术通报, 2010(2): 76-84. | |
| 30 | Magali L, Patrice D, Gert T, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 2002, 30(1): 325-327. |
| 31 | Benedito V A, Torres-Jerez I, Murray J D, et al. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal, 2008, 55(3): 504-513. |
| 32 | Meng C M, Ji J H, Li X L, et al. Electronic clone and bioinformatics analysis of CPP transcription factor genes from wheat. Biotechnology, 2014, 24(4): 39-42. |
| 孟超敏, 姬俊华, 李雪林, 等. 小麦CPP转录因子基因的电子克隆及生物信息学分析. 生物技术, 2014, 24(4): 39-42. | |
| 33 | Goh C S, Bogan A A, Joachimiak M, et al. Co-evolution of proteins with their interaction partners. Journal of Molecular Biology, 2000, 299(2): 283-293. |
| 34 | Jain M, Tyagi A K, Khurana J P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics, 2006, 88(3): 360-371. |
| 35 | Zhang M J, Zhu L, Xia Q Z. Research progress on the regulation of plant hormones to stress responses. Journal of Hubei University (Natural Science), 2021, 43(3): 242-253, 263. |
| 张明菊, 朱莉, 夏启中. 植物激素对胁迫反应调控的研究进展. 湖北大学学报(自然科学版), 2021, 43(3): 242-253, 263. | |
| 36 | Pan R R, Wei M M, Wang Y J, et al. Cloning and expression analysis of HbCCP1 in rubber tree (Hevea brasiliensis). Plant Physiology Journal, 2018, 54(5): 763-772. |
| 潘冉冉, 位明明, 王亚杰, 等. 巴西橡胶树HbCPP1基因的克隆与表达分析. 植物生理学报, 2018, 54(5): 763-772. |
| [1] | 刘亚男, 于人杰, 高燕丽, 康俊梅, 杨青川, 武志海, 王珍. 蒺藜苜蓿膜联蛋白MtANN2基因的表达模式及盐胁迫下的功能分析[J]. 草业学报, 2022, 31(5): 124-134. |
| [2] | 高莉娟, 张正社, 文裕, 宗西方, 闫启, 卢丽燕, 易显凤, 张吉宇. 象草全基因组bHLH转录因子家族鉴定及表达分析[J]. 草业学报, 2022, 31(3): 47-59. |
| [3] | 魏娜, 李艳鹏, 马艺桐, 刘文献. 全基因组水平紫花苜蓿TCP基因家族的鉴定及其在干旱胁迫下表达模式分析[J]. 草业学报, 2022, 31(1): 118-130. |
| [4] | 杨志民, 邢瑞, 丁鋆嘉, 庄黎丽. 基于转录组测序的高羊茅分蘖与株高相关差异表达基因分析[J]. 草业学报, 2022, 31(1): 145-163. |
| [5] | 张家驹, 于洁, 李明娜, 康俊梅, 杨青川, 龙瑞才. 蒺藜苜蓿lncRNA167及其剪切产物miR167c的鉴定和功能分析[J]. 草业学报, 2022, 31(1): 164-180. |
| [6] | 吴彤, 刘云苗, 金军, 董伟峰, 才晓溪, 孙明哲, 贾博为, 孙晓丽. 蒺藜苜蓿cation/H+ exchanger基因家族鉴定及表达特征分析[J]. 草业学报, 2022, 31(1): 181-194. |
| [7] | 周晶, 陈思齐, 史文娇, 阳伏林, 林辉, 林占熺. 巨菌草幼叶及根转录组功能基因测序及分析[J]. 草业学报, 2021, 30(2): 143-155. |
| [8] | 王如月, 文武武, 赵恩华, 周鹏, 安渊. 紫花苜蓿MsWRKY11基因的克隆及其耐盐功能分析[J]. 草业学报, 2021, 30(11): 157-169. |
| [9] | 侯洁茹, 段晓玥, 李州, 彭燕. 白三叶TrSAMDC1克隆及表达分析[J]. 草业学报, 2020, 29(8): 170-178. |
| [10] | 罗维, 舒健虹, 刘晓霞, 王子苑, 牟琼, 王小利, 吴佳海. 高羊茅FaRVE8基因的克隆、亚细胞定位及表达分析[J]. 草业学报, 2020, 29(7): 60-69. |
| [11] | 刘文文, 崔会婷, 尉春雪, 龙瑞才, 康俊梅, 杨青川, 王珍. 蒺藜苜蓿叶绿素酸酯a加氧酶(MtCAO)基因的克隆与功能分析[J]. 草业学报, 2020, 29(5): 171-181. |
| [12] | 杨婷, 张建平, 刘自刚, 齐燕妮, 李闻娟, 谢亚萍. 胡麻异质型ACCase亚基基因的克隆与表达分析[J]. 草业学报, 2020, 29(4): 111-120. |
| [13] | 黄沁梅, 杨伊如, 陈丽飞, 尹航, 刘贺, 刘颖婕, 周蕴薇, 何淼. 转露地菊CgDREB22基因的烟草抗逆性分析[J]. 草业学报, 2020, 29(10): 109-118. |
| [14] | 杨柳慧, 尹航, 黄沁梅, 张彦妮, 何淼, 周蕴薇. 细叶百合LpWRKY20基因对非生物胁迫的响应及抗旱性分析[J]. 草业学报, 2020, 29(1): 193-202. |
| [15] | 夏曾润, 王文颖, 刘亚琪, 王锁民. 罗布麻K+通道编码基因AvAKT1的克隆与表达分析[J]. 草业学报, 2019, 28(8): 180-189. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||