

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (1): 118-130.DOI: 10.11686/cyxb2021189
• 研究论文 • 上一篇
收稿日期:2021-05-07
修回日期:2021-07-07
出版日期:2021-12-01
发布日期:2021-12-01
通讯作者:
刘文献
作者简介:Corresponding author. E-mail: liuwx@lzu.edu.cn基金资助:
Na WEI( ), Yan-peng LI, Yi-tong MA, Wen-xian LIU(
), Yan-peng LI, Yi-tong MA, Wen-xian LIU( )
)
Received:2021-05-07
Revised:2021-07-07
Online:2021-12-01
Published:2021-12-01
Contact:
Wen-xian LIU
摘要:
紫花苜蓿是世界最重要的豆科牧草之一,干旱是影响其产量和地理分布的关键瓶颈。在紫花苜蓿响应干旱胁迫过程中,转录因子发挥着重要的调控作用。TCP(teosinte branchesd 1/cycloidea/pro-liferating cell factors)为植物特有的转录因子,在植物生长、发育、响应逆境胁迫中都具有重要的生物学功能。截至目前,该基因家族在紫花苜蓿中的分布以及响应干旱胁迫的生物学功能仍未见报道。因此,为进一步挖掘紫花苜蓿中响应干旱胁迫功能基因,本研究利用生物信息学方法在全基因组水平对TCP基因家族进行了鉴定,并对其系统进化、基因结构、染色体定位、共线性分析以及干旱胁迫下的表达模式进行了分析。结果表明,紫花苜蓿基因组中共鉴定出40个MsTCP基因,不均匀地分布于20条染色体上,其中包括17对旁系同源基因对,且都是基因片段复制事件。系统发育和保守结构域分析发现,MsTCP基因可以分为2个大分支和3个亚家族(PCF, CIN与CYC/TB1),同一分支中的成员具有相同氨基酸数目的TCP结构域,同亚家族中的成员具有相似的保守基序与基因结构。此外,通过分析紫花苜蓿响应干旱转录组数据共鉴定出4个可能与紫花苜蓿响应干旱胁迫有关的MsTCP基因(MsTCP23,MsTCP27,MsTCP29,MsTCP33)。qRT-PCR结果进一步表明PEG模拟干旱胁迫处理后,这4个基因的表达量在根和叶中均显著上调,进一步确定了这些基因的确响应紫花苜蓿干旱胁迫。该研究为后期深入解析紫花苜蓿响应干旱胁迫理论以及通过基因工程技术创制高抗旱紫花苜蓿新种质奠定基础。
魏娜, 李艳鹏, 马艺桐, 刘文献. 全基因组水平紫花苜蓿TCP基因家族的鉴定及其在干旱胁迫下表达模式分析[J]. 草业学报, 2022, 31(1): 118-130.
Na WEI, Yan-peng LI, Yi-tong MA, Wen-xian LIU. Genome-wide identification of the alfalfa TCP gene family and analysis of gene transcription patterns in alfalfa (Medicago sativa) under drought stress[J]. Acta Prataculturae Sinica, 2022, 31(1): 118-130.
| 引物名称Primer name | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsTCP23 | CAAGATATGACAATGACAGTGCC | GAACCTTAAACCTTCCTTCCTC |
| MsTCP27 | GTTACAAAGACCAAATCACCCT | AAGCCATTGTTTCCCAATTGAG |
| MsTCP29 | CAAGATATGACAATGACAGTGCC | CTTCCTTCCTTTGACACGAC |
| MsTCP33 | AACCCTAATCAAGAACCAAACC | GTATAGCCCATACAGGAACCA |
| MsActin | GACAATGGAACTGGAATGG | CAATACCGTGCTCAATGG |
表1 引物信息及序列
Table 1 The information of the primer and sequences used in this study
| 引物名称Primer name | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsTCP23 | CAAGATATGACAATGACAGTGCC | GAACCTTAAACCTTCCTTCCTC |
| MsTCP27 | GTTACAAAGACCAAATCACCCT | AAGCCATTGTTTCCCAATTGAG |
| MsTCP29 | CAAGATATGACAATGACAGTGCC | CTTCCTTCCTTTGACACGAC |
| MsTCP33 | AACCCTAATCAAGAACCAAACC | GTATAGCCCATACAGGAACCA |
| MsActin | GACAATGGAACTGGAATGG | CAATACCGTGCTCAATGG |
基因名 Gene name | 基因ID Gene ID | 蛋白长度 Protein length (aa) | 分子量 Molecular weight (Da) | 等电点 pI | 亚组 Subgroup | 蛋白亲水性 Protein GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| MsTCP1 | MS.gene95781.t1 | 421 | 47569.61 | 6.43 | CYC/TB1 | -0.764 | nucl |
| MsTCP2 | MS.gene073917.t1 | 143 | 16100.50 | 11.55 | PCF | -0.535 | nucl |
| MsTCP3 | MS.gene34439.t1 | 326 | 34413.31 | 4.84 | PCF | -0.351 | nucl |
| MsTCP4 | MS.gene029214.t1 | 260 | 27872.04 | 9.72 | PCF | -0.575 | nucl |
| MsTCP5 | MS.gene91110.t1 | 232 | 25464.17 | 8.04 | PCF | -0.819 | nucl |
| MsTCP6 | MS.gene41379.t1 | 340 | 35 855.73 | 4.85 | PCF | -0.386 | nucl |
| MsTCP7 | MS.gene88588.t1 | 238 | 26275.98 | 8.05 | PCF | -0.889 | nucl |
| MsTCP8 | MS.gene064482.t1 | 206 | 22076.72 | 8.96 | PCF | -0.527 | nucl |
| MsTCP9 | MS.gene060651.t1 | 300 | 32841.10 | 7.14 | CIN | -0.739 | nucl |
| MsTCP10 | MS.gene31403.t1 | 329 | 36285.54 | 5.99 | CIN | -0.743 | nucl |
| MsTCP11 | MS.gene059738.t1 | 302 | 33141.45 | 7.13 | CIN | -0.731 | nucl |
| MsTCP12 | MS.gene072060.t1 | 520 | 55104.71 | 6.32 | PCF | -0.782 | nucl |
| MsTCP13 | MS.gene93507.t1 | 383 | 43316.86 | 7.92 | CIN | -0.799 | nucl |
| MsTCP14 | MS.gene006670.t1 | 336 | 37829.29 | 6.21 | CIN | -0.881 | nucl |
| MsTCP15 | MS.gene023326.t1 | 478 | 53008.41 | 6.75 | CIN | -0.978 | nucl |
| MsTCP16 | MS.gene73844.t1 | 224 | 24733.21 | 7.23 | CIN | -0.795 | nucl |
| MsTCP17 | MS.gene78508.t1 | 417 | 45864.22 | 8.02 | PCF | -0.607 | nucl |
| MsTCP18 | MS.gene91902.t1 | 415 | 43919.25 | 6.46 | PCF | -0.613 | nucl |
| MsTCP19 | MS.gene054307.t1 | 119 | 12818.73 | 9.59 | PCF | -0.576 | nucl |
| MsTCP20 | MS.gene054308.t1 | 134 | 14873.93 | 7.76 | PCF | -0.704 | nucl |
| MsTCP21 | MS.gene054297.t1 | 119 | 12797.68 | 10.00 | PCF | -0.612 | nucl |
| MsTCP22 | MS.gene03516.t1 | 120 | 12812.73 | 9.88 | PCF | -0.549 | nucl |
| MsTCP23 | MS.gene03512.t1 | 134 | 14658.83 | 8.82 | PCF | -0.476 | nucl |
| MsTCP24 | MS.gene074319.t1 | 319 | 36194.52 | 9.67 | CYC/TB1 | -1.033 | nucl |
| MsTCP25 | MS.gene80495.t1 | 107 | 11418.15 | 10.23 | PCF | -0.508 | nucl |
| MsTCP26 | MS.gene80502.t1 | 164 | 18081.65 | 7.72 | PCF | -0.476 | nucl |
| MsTCP27 | MS.gene48573.t1 | 351 | 39672.79 | 8.57 | CIN | -0.944 | nucl |
| MsTCP28 | MS.gene063503.t1 | 309 | 34369.17 | 9.10 | CIN | -0.678 | nucl |
| MsTCP29 | MS.gene42033.t1 | 130 | 14327.45 | 9.46 | PCF | -0.555 | cyto |
| MsTCP30 | MS.gene42025.t1 | 120 | 12784.56 | 10.14 | PCF | -0.653 | nucl |
| MsTCP31 | MS.gene92019.t1 | 373 | 42567.64 | 6.68 | CYC/TB1 | -1.135 | nucl |
| MsTCP32 | MS.gene36024.t1 | 435 | 47331.52 | 6.73 | CIN | -0.831 | nucl |
| MsTCP33 | MS.gene019430.t1 | 331 | 36115.04 | 9.42 | PCF | -0.447 | nucl |
| MsTCP34 | MS.gene044458.t1 | 249 | 27766.24 | 9.25 | CIN | -0.668 | nucl |
| MsTCP35 | MS.gene36926.t1 | 189 | 20950.04 | 9.55 | PCF | -0.708 | cyto |
| MsTCP36 | MS.gene032256.t1 | 433 | 47041.11 | 6.59 | CIN | -0.839 | nucl |
| MsTCP37 | MS.gene033131.t1 | 419 | 45864.22 | 8.02 | PCF | -0.895 | nucl |
| MsTCP38 | MS.gene007917.t1 | 431 | 46871.96 | 6.61 | CIN | -0.823 | nucl |
| MsTCP39 | MS.gene038752.t1 | 388 | 44616.54 | 8.50 | CYC/TB1 | -1.012 | nucl |
| MsTCP40 | MS.gene34255.t1 | 430 | 46850.01 | 6.71 | CIN | -0.834 | nucl |
表2 紫花苜蓿TCP家族成员基本信息
Table 2 Basic information of TCP family members in alfalfa
基因名 Gene name | 基因ID Gene ID | 蛋白长度 Protein length (aa) | 分子量 Molecular weight (Da) | 等电点 pI | 亚组 Subgroup | 蛋白亲水性 Protein GRAVY | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| MsTCP1 | MS.gene95781.t1 | 421 | 47569.61 | 6.43 | CYC/TB1 | -0.764 | nucl |
| MsTCP2 | MS.gene073917.t1 | 143 | 16100.50 | 11.55 | PCF | -0.535 | nucl |
| MsTCP3 | MS.gene34439.t1 | 326 | 34413.31 | 4.84 | PCF | -0.351 | nucl |
| MsTCP4 | MS.gene029214.t1 | 260 | 27872.04 | 9.72 | PCF | -0.575 | nucl |
| MsTCP5 | MS.gene91110.t1 | 232 | 25464.17 | 8.04 | PCF | -0.819 | nucl |
| MsTCP6 | MS.gene41379.t1 | 340 | 35 855.73 | 4.85 | PCF | -0.386 | nucl |
| MsTCP7 | MS.gene88588.t1 | 238 | 26275.98 | 8.05 | PCF | -0.889 | nucl |
| MsTCP8 | MS.gene064482.t1 | 206 | 22076.72 | 8.96 | PCF | -0.527 | nucl |
| MsTCP9 | MS.gene060651.t1 | 300 | 32841.10 | 7.14 | CIN | -0.739 | nucl |
| MsTCP10 | MS.gene31403.t1 | 329 | 36285.54 | 5.99 | CIN | -0.743 | nucl |
| MsTCP11 | MS.gene059738.t1 | 302 | 33141.45 | 7.13 | CIN | -0.731 | nucl |
| MsTCP12 | MS.gene072060.t1 | 520 | 55104.71 | 6.32 | PCF | -0.782 | nucl |
| MsTCP13 | MS.gene93507.t1 | 383 | 43316.86 | 7.92 | CIN | -0.799 | nucl |
| MsTCP14 | MS.gene006670.t1 | 336 | 37829.29 | 6.21 | CIN | -0.881 | nucl |
| MsTCP15 | MS.gene023326.t1 | 478 | 53008.41 | 6.75 | CIN | -0.978 | nucl |
| MsTCP16 | MS.gene73844.t1 | 224 | 24733.21 | 7.23 | CIN | -0.795 | nucl |
| MsTCP17 | MS.gene78508.t1 | 417 | 45864.22 | 8.02 | PCF | -0.607 | nucl |
| MsTCP18 | MS.gene91902.t1 | 415 | 43919.25 | 6.46 | PCF | -0.613 | nucl |
| MsTCP19 | MS.gene054307.t1 | 119 | 12818.73 | 9.59 | PCF | -0.576 | nucl |
| MsTCP20 | MS.gene054308.t1 | 134 | 14873.93 | 7.76 | PCF | -0.704 | nucl |
| MsTCP21 | MS.gene054297.t1 | 119 | 12797.68 | 10.00 | PCF | -0.612 | nucl |
| MsTCP22 | MS.gene03516.t1 | 120 | 12812.73 | 9.88 | PCF | -0.549 | nucl |
| MsTCP23 | MS.gene03512.t1 | 134 | 14658.83 | 8.82 | PCF | -0.476 | nucl |
| MsTCP24 | MS.gene074319.t1 | 319 | 36194.52 | 9.67 | CYC/TB1 | -1.033 | nucl |
| MsTCP25 | MS.gene80495.t1 | 107 | 11418.15 | 10.23 | PCF | -0.508 | nucl |
| MsTCP26 | MS.gene80502.t1 | 164 | 18081.65 | 7.72 | PCF | -0.476 | nucl |
| MsTCP27 | MS.gene48573.t1 | 351 | 39672.79 | 8.57 | CIN | -0.944 | nucl |
| MsTCP28 | MS.gene063503.t1 | 309 | 34369.17 | 9.10 | CIN | -0.678 | nucl |
| MsTCP29 | MS.gene42033.t1 | 130 | 14327.45 | 9.46 | PCF | -0.555 | cyto |
| MsTCP30 | MS.gene42025.t1 | 120 | 12784.56 | 10.14 | PCF | -0.653 | nucl |
| MsTCP31 | MS.gene92019.t1 | 373 | 42567.64 | 6.68 | CYC/TB1 | -1.135 | nucl |
| MsTCP32 | MS.gene36024.t1 | 435 | 47331.52 | 6.73 | CIN | -0.831 | nucl |
| MsTCP33 | MS.gene019430.t1 | 331 | 36115.04 | 9.42 | PCF | -0.447 | nucl |
| MsTCP34 | MS.gene044458.t1 | 249 | 27766.24 | 9.25 | CIN | -0.668 | nucl |
| MsTCP35 | MS.gene36926.t1 | 189 | 20950.04 | 9.55 | PCF | -0.708 | cyto |
| MsTCP36 | MS.gene032256.t1 | 433 | 47041.11 | 6.59 | CIN | -0.839 | nucl |
| MsTCP37 | MS.gene033131.t1 | 419 | 45864.22 | 8.02 | PCF | -0.895 | nucl |
| MsTCP38 | MS.gene007917.t1 | 431 | 46871.96 | 6.61 | CIN | -0.823 | nucl |
| MsTCP39 | MS.gene038752.t1 | 388 | 44616.54 | 8.50 | CYC/TB1 | -1.012 | nucl |
| MsTCP40 | MS.gene34255.t1 | 430 | 46850.01 | 6.71 | CIN | -0.834 | nucl |
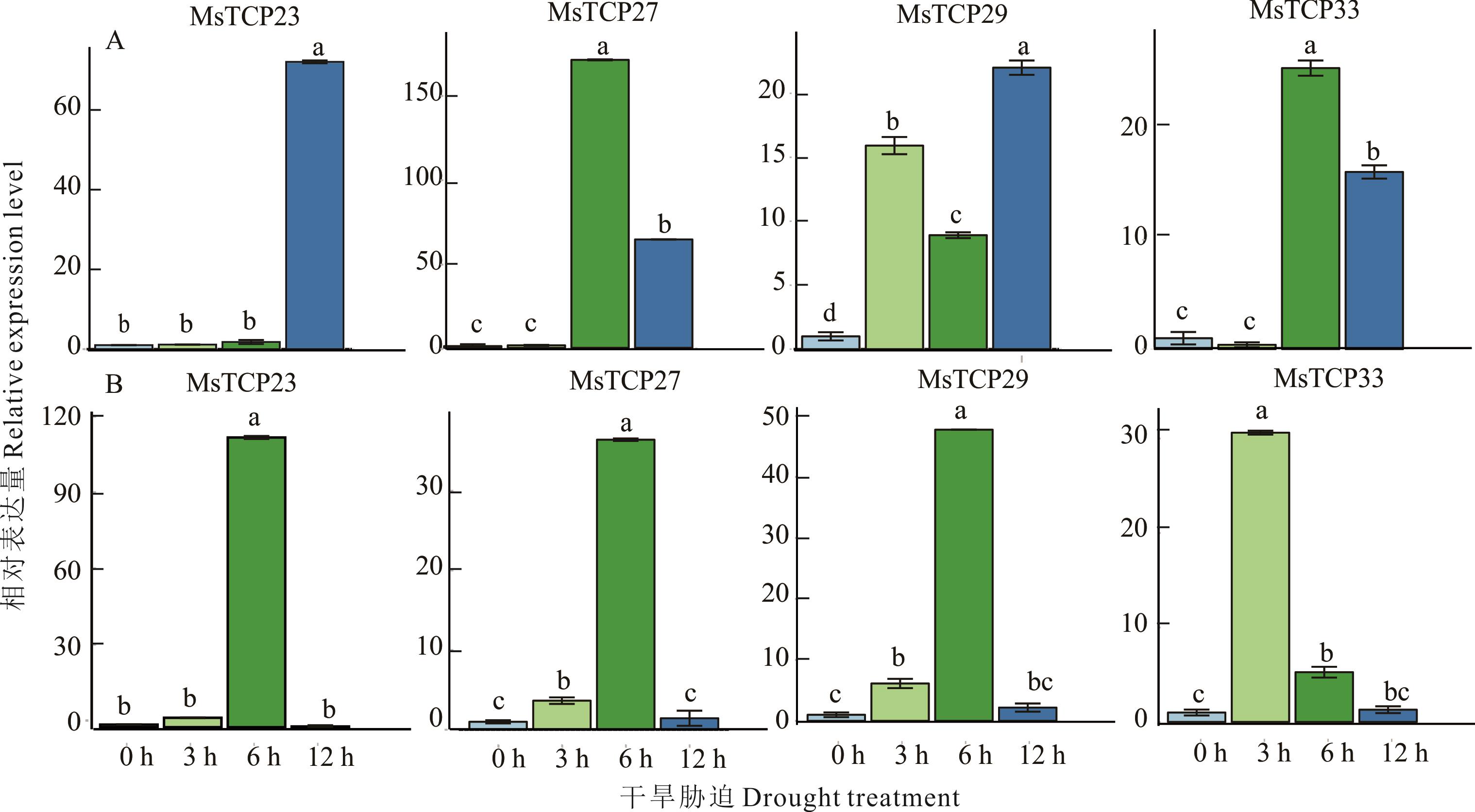
图8 MsTCP基因响应干旱胁迫的表达模式A:叶片;B:根。数据是三个重复的平均值,不同小写字母表示0.05水平存在显著性差异。A: Leaf; B: Root. The data is the average of three repetitions, the differernt lowercase letters indicate significant differences at the 0.05 level.
Fig.8 Expression patterns of MsTCP genes in response to drought stress
| 1 | Almeida D M, Gregorio G B, Oliveira M M, et al. Five novel transcription factors as potential regulators of osnhx1 gene expression in a salt tolerant rice genotype. Plant Molecular Biology, 2016, 93(1/2): 1-17. |
| 2 | Cubas P, Lauter N, Doebley J, et al. The TCP domain: a motif found in proteins regulating plant growth and development. Plant Journal, 2010, 18(2): 215-222. |
| 3 | Doebley J, Stec A. Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 1995, 141(1): 333-346. |
| 4 | Luo D, Carpenter R, Vincent C, et al. Origin of floral asymmetry in Antirrhinum. Nature, 1996, 383(6603): 794-799. |
| 5 | Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. The Plant Cell, 1997, 9(9): 1607-1619. |
| 6 | Navaud O, Dabos P, Carnus E, et al. TCP transcription factors predate the emergence of land plants. Journal of Molecular Evolution, 2007, 65(1): 23-33. |
| 7 | Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends in Plant Science, 2010, 15(1): 31-39. |
| 8 | Yao X, Hong M, Jian W, et al. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. Journal of Integrative Plant Biology, 2007, 49(6): 885-897. |
| 9 | Huo Y, Xiong W, Su K, et al. Genome-wide analysis of the TCP gene family in Switchgrass (Panicum virgatum L.). International Journal of Genomics, 2019(1): 1-13. |
| 10 | Parapunova V, Busscher M, Busscher-Lange J, et al. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biology, 2014, 14(1): 1-17. |
| 11 | Danisman S, Wal F, Dhondt S, et al. Arabidopsis class I and class Ⅱ TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiology, 2012, 159(4): 1511-1523. |
| 12 | Wang H F, Wang H W, Liu R, et al. Genome-wide identification of TCP family transcription factors in Medicago truncatula reveals significant roles of miR319-Targeted TCPs in nodule development. Frontiers in Plant Science, 2018, 9: 774. |
| 13 | Sharma R, Kapoor M, Tyagi A K, et al. Comparative transcript profiling of TCP family genes provide insight into gene functions and diversification in rice and Arabidopsis. Journal of Plant Molecular Biology & Biotechnology, 2010, 1(1): 24-38. |
| 14 | Mukhopadhyay P, Tyagi A K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Scientific Reports, 2015, 5(1): 9998. |
| 15 | Liu C H, Liang N S, Yu L, et al. Fraxinus mandshurica Rupr. TCP4 transcription factor cloning and expression analysis under stress and hormones. Journal of Beijing Forestry University, 2017,39(6): 22-31. |
| 刘春浩, 梁楠松, 于磊, 等. 水曲柳TCP4转录因子克隆及胁迫和激素下的表达分析. 北京林业大学学报, 2017, 39(6): 22-31. | |
| 16 | Lei N, Li S X, Peng M. Cloning, expression analysis and plant expression vector construction of cassava MeTCP4 transcription factor. Molecular Plant Breeding, 2018, 16(5): 1517-1523. |
| 雷宁, 李淑霞, 彭明. 木薯MeTCP4转录因子的克隆, 表达分析及植物表达载体的构建. 分子植物育种, 2018, 16(5): 1517-1523. | |
| 17 | Liu Z, Chen T, Ma L, et al. Global transcriptome sequencing using the illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS One, 2013, 8(12): e83549. |
| 18 | Tang L, Cai H, Zhai H, et al. Overexpression of Glycine sojaWRKY20 enhances both drought and salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Cell, Tissue and Organ Culture, 2014, 118(1): 77-86. |
| 19 | Jin X, Yin X, Ndayambaza B, et al. Genome-wide identification and expression profiling of the ERF gene family in Medicago sativa L. under various abiotic stresses. DNA and Cell Biology, 2019, 38(10): 1056-1068. |
| 20 | Min X, Jin X, Zhang Z, et al. Genome-wide identification of NAC transcription factor family and functional analysis of the abiotic stress-responsive genes in Medicago sativa L.Journal of Plant Growth Regulation, 2020, 39(1): 324-337. |
| 21 | Chen H, Zeng Y, Yang Y, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nature Communications, 2020, 11(1): 1-11. |
| 22 | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 2012, 40: 597-603. |
| 23 | Sara E G, Jaina M, Alex B, et al. The Pfam protein families database in 2019. Nucleic Acids Research, 2019,47(D1): 427-432. |
| 24 | Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics, 1994, 10(2): 189-191. |
| 25 | Yuan J, Amend A, Borkowski J, et al. MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics, 1999, 15(10): 862-863. |
| 26 | Hall T, Biosciences I, Carlsbad C. BioEdit: an important software for molecular biology. GERF Bulletin Biosciences, 2011, 2(1): 60-61. |
| 27 | Bailey T L, Boden M, Buske F A, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research, 2009, 37(2): W202-W208. |
| 28 | Chen C, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 29 | Wang Y, Tang H, Debarry J D, et al. MCScanX a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 2012, 40(7): e49. |
| 30 | Luo D, Wu Y, Liu J, et al. Comparative transcriptomic and physiological analyses of Medicago sativa L. indicates that multiple regulatory networks are activated during continuous ABA treatment. International Journal of Molecular Sciences, 2018, 20(1): 47. |
| 31 | Min X, Liu Z, Wang Y, et al. Comparative transcriptomic analysis provides insights into the coordinated mechanisms of leaves and roots response to cold stress in Common Vetch. Industrial Crops and Products, 2020, 158: 112949. |
| 32 | Nei M. Gene duplication and nucleotide substitution in evolution. Nature, 1969, 221(5175): 40-42. |
| 33 | Zheng L, Bai X T, Li H Y. Genome-wide identification and expression analysis of Sorghum bicolorTCP gene family. Henan Agricultural Sciences, 2019, 537(10): 36-42. |
| 郑玲, 白雪婷, 李会云. 高粱TCP基因家族全基因组鉴定及表达分析. 河南农业科学, 2019, 537(10): 36-42. | |
| 34 | Liu Y, Zhang H, Xin D W, et al. Analysis and function prediction of Glycine maxTCP transcription factor domains. Soybean Science, 2012, 31(5): 707-713. |
| 刘洋, 张慧, 辛大伟, 等. 大豆TCP转录因子家族结构域分析及功能预测. 大豆科学, 2012, 31(5): 707-713. | |
| 35 | Li K J, Tan S S, Sun B, et al. Genome-wide identification and expression analysis of Brassica juncea TCP transcription factor. Journal of Sichuan Agricultural University, 2019, 37(4): 459-468. |
| 李坤杰, 谭杉杉, 孙勃, 等. 芥菜TCP转录因子家族全基因组鉴定及表达分析. 四川农业大学学报, 2019, 37(4): 459-468. | |
| 36 | Lei D, Wu Y, Su Z, et al. Research progress on the interaction between TCP transcription factors and hormone signals. Molecular Plant Breeding, 2019, 17(9): 2868-2875. |
| 雷豆, 吴雨, 苏周, 等. TCP转录因子与激素信号相互作用研究进展. 分子植物育种, 2019, 17(9): 2868-2875. | |
| 37 | Sun H D, Xue J Z, Liu Y J, et al. Genome-wide identification and expression pattern analysis of maize TCP transcription factor family. Molecular Plant Breeding, 2021, 19(8): 2460-2471. |
| 孙菡笛, 薛江芝, 刘亚洁, 等. 玉米 TCP 转录因子家族的全基因组鉴定及表达模式分析. 分子植物育种, 2021, 19(8): 2460-2471. | |
| 38 | Perez M, Guerringue Y, Ranty B, et al. Specific TCP transcription factors interact with and stabilize PRR2 within different nuclear sub-domains. Plant Science, 2019, 287: 110197. |
| 39 | Sun X, Wang C, Xiang N, et al. Activation of secondary cell wall biosynthesis by miR319‐targeted TCP4 transcription factor. Plant Biotechnology Journal, 2017, 15(10): 1284-1294. |
| 40 | Ma X, Ma J, Fan D, et al. Genome-wide identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Scientific Reports, 2016, 6: 32795. |
| 41 | Zhao M, Peng X, Chen N, et al. Genome-wide identification of the TCP gene family in Broussonetia papyrifera and functional analysis of BpTCP8, 14 and 19 in shoot branching. Plants, 2020, 9(10): 1301. |
| 42 | Leng X, Wei H, Xu X, et al. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics, 2019, 20(1): 1-18. |
| 43 | Liu Y, Guan X, Liu S, et al. Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. International Journal of Molecular Sciences, 2018, 19(3): 847. |
| [1] | 赵颖, 辛夏青, 魏小红. 一氧化氮对干旱胁迫下紫花苜蓿氮代谢的影响[J]. 草业学报, 2021, 30(9): 86-96. |
| [2] | 汪雪, 刘晓静, 赵雅姣, 王静. 根系分隔方式下紫花苜蓿/燕麦间作氮素利用及种间互馈特征研究[J]. 草业学报, 2021, 30(8): 73-85. |
| [3] | 古丽娜扎尔·艾力null, 陶海宁, 王自奎, 沈禹颖. 基于APSIM模型的黄土旱塬区苜蓿——小麦轮作系统深层土壤水分及水分利用效率研究[J]. 草业学报, 2021, 30(7): 22-33. |
| [4] | 周倩倩, 张亚见, 张静, 殷涂童, 盛下放, 何琳燕. 产硫化氢细菌的筛选及阻控苜蓿吸收铅和改良土壤的作用[J]. 草业学报, 2021, 30(7): 44-52. |
| [5] | 臧真凤, 白婕, 刘丛, 昝看卓, 龙明秀, 何树斌. 紫花苜蓿形态和生理指标响应干旱胁迫的品种特异性[J]. 草业学报, 2021, 30(6): 73-81. |
| [6] | 谢展, 穆麟, 张志飞, 陈桂华, 刘洋, 高帅, 魏仲珊. 乳酸菌或有机酸盐与尿素复配添加对紫花苜蓿混合青贮的影响[J]. 草业学报, 2021, 30(5): 165-173. |
| [7] | 王吉祥, 宫焕宇, 屠祥建, 郭侲洐, 赵嘉楠, 沈健, 栗振义, 孙娟. 耐亚磷酸盐紫花苜蓿品种筛选及评价指标的鉴定[J]. 草业学报, 2021, 30(5): 186-199. |
| [8] | 罗巧玉, 王彦龙, 陈志, 马永贵, 任启梅, 马玉寿. 水分逆境对发草脯氨酸及其代谢途径的影响[J]. 草业学报, 2021, 30(5): 75-83. |
| [9] | 张小芳, 魏小红, 刘放, 朱雪妹. PEG胁迫下紫花苜蓿幼苗内源激素对NO的响应[J]. 草业学报, 2021, 30(4): 160-169. |
| [10] | 候怡谣, 李霄, 龙瑞才, 杨青川, 康俊梅, 郭长虹. 过量表达紫花苜蓿MsHB7基因对拟南芥耐旱性的影响[J]. 草业学报, 2021, 30(4): 170-179. |
| [11] | 刘凯强, 刘文辉, 贾志锋, 梁国玲, 马祥. 干旱胁迫对‘青燕1号’燕麦产量及干物质积累与分配的影响[J]. 草业学报, 2021, 30(3): 177-188. |
| [12] | 马欣, 罗珠珠, 张耀全, 刘家鹤, 牛伊宁, 蔡立群. 黄土高原雨养区不同种植年限紫花苜蓿土壤细菌群落特征与生态功能预测[J]. 草业学报, 2021, 30(3): 54-67. |
| [13] | 沙栢平, 谢应忠, 高雪芹, 蔡伟, 伏兵哲. 地下滴灌水肥耦合对紫花苜蓿草产量及品质的影响[J]. 草业学报, 2021, 30(2): 102-114. |
| [14] | 刘晓静, 赵雅姣, 郝凤, 童长春. 紫花苜蓿氮效率及其类型特征研究[J]. 草业学报, 2021, 30(12): 90-102. |
| [15] | 马倩, 闫启, 张正社, 吴凡, 张吉宇. 紫花苜蓿CCoAOMT基因家族的鉴定、进化及表达分析[J]. 草业学报, 2021, 30(11): 144-156. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||