

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (3): 47-59.DOI: 10.11686/cyxb2020593
高莉娟1( ), 张正社1, 文裕2, 宗西方1, 闫启1, 卢丽燕1, 易显凤3, 张吉宇1(
), 张正社1, 文裕2, 宗西方1, 闫启1, 卢丽燕1, 易显凤3, 张吉宇1( )
)
收稿日期:2020-12-29
修回日期:2021-02-09
出版日期:2022-03-20
发布日期:2022-01-15
通讯作者:
张吉宇
作者简介:Corresponding author. E-mail: zhangjy@lzu.edu.cn基金资助:
Li-juan GAO1( ), Zheng-she ZHANG1, Yu WEN2, Xi-fang ZONG1, Qi YAN1, Li-yan LU1, Xian-feng YI3, Ji-yu ZHANG1(
), Zheng-she ZHANG1, Yu WEN2, Xi-fang ZONG1, Qi YAN1, Li-yan LU1, Xian-feng YI3, Ji-yu ZHANG1( )
)
Received:2020-12-29
Revised:2021-02-09
Online:2022-03-20
Published:2022-01-15
Contact:
Ji-yu ZHANG
摘要:
bHLH转录因子家族不仅参与了植物的生长发育,而且在植物响应逆境胁迫和次生代谢方面发挥着关键作用。在全基因组水平对重要饲草及能源植物象草的bHLH转录因子家族进行了鉴定及分析,并利用转录组数据及定量RT-PCR分析了象草bHLH转录因子对赤霉素(GA3)和多效唑(PAC)的响应。结果显示:在象草中共鉴定出229个具有完整保守结构域的bHLH基因家族成员(CpbHLH001~CpbHLH229),不均匀地分布于14条染色体上;系统进化分析结果表明,229个CpbHLHs可被分为18个亚类,其中C亚类的成员数量最多,为41个;此外,相同亚家族中的大多数基因具有相似的基因结构和保守基序;基于转录组数据的表达谱分析结果发现,多数bHLH基因在象草茎尖组织中均对GA3和PAC有响应。随机挑选9个表达量较高的基因进一步通过qPCR进行验证。结果显示,经外源GA3和PAC处理之后,这9个基因因不同处理而差异表达,表明这9个基因可能与GA3和PAC介导的信号通路有关。综上所述,本研究为象草bHLH转录因子家族的生物学功能奠定了基础。
高莉娟, 张正社, 文裕, 宗西方, 闫启, 卢丽燕, 易显凤, 张吉宇. 象草全基因组bHLH转录因子家族鉴定及表达分析[J]. 草业学报, 2022, 31(3): 47-59.
Li-juan GAO, Zheng-she ZHANG, Yu WEN, Xi-fang ZONG, Qi YAN, Li-yan LU, Xian-feng YI, Ji-yu ZHANG. Genome-wide identification and expression analysis of the bHLH transcription factor family in Cenchrus purpureus[J]. Acta Prataculturae Sinica, 2022, 31(3): 47-59.
| 基因名称Gene name | 上游引物Forward primer (5'-3') | 下游引物Reverse primer (5'-3') |
|---|---|---|
| CpbHLH002 | CAAGATCAGCAAGATGGACAGG | GGGTGACTCAAGCTCATTCTG |
| CpbHLH013 | ACAAGCAAACCAACACTTCTG | GCTCTCCTTTAACACCTTAACCT |
| CpbHLH017 | AGCAAGTTGAGTTCTTGTCC | TCTTTGTGCAGGAGTGTAGAC |
| CpbHLH019 | TGCGGAAAGGTTGAGAAGG | TGATCTCGTCAAGCATCACC |
| CpbHLH025 | ACAAACAAGACAGATAGGGCAG | GAGAGTGGAATATCAGCAACCAG |
| CpbHLH030 | ACAAGCAAACCAACACTTCAG | CTCGTTCAGCACCTTGACC |
| CpbHLH055 | GTCAAGGTACTAAGCATGAGCA | TCTTCTTCCATCAGCTTTGTG |
| CpbHLH156 | AGCAACAGCGAGAATGGC | CAGCAGCTTCATCCTCTGG |
| CpbHLH158 | AGCTGGTTGCTGATATTCCAC | TCTTCCATCAGCTTTGCAACC |
| CpEE1a | ACTACGAGCAAGAGTTGGAAAC | TGGATGGCTGGAAGAGGAC |
表1 实时定量PCR引物序列
Table 1 Primer sequences used in real-time quantitative PCR
| 基因名称Gene name | 上游引物Forward primer (5'-3') | 下游引物Reverse primer (5'-3') |
|---|---|---|
| CpbHLH002 | CAAGATCAGCAAGATGGACAGG | GGGTGACTCAAGCTCATTCTG |
| CpbHLH013 | ACAAGCAAACCAACACTTCTG | GCTCTCCTTTAACACCTTAACCT |
| CpbHLH017 | AGCAAGTTGAGTTCTTGTCC | TCTTTGTGCAGGAGTGTAGAC |
| CpbHLH019 | TGCGGAAAGGTTGAGAAGG | TGATCTCGTCAAGCATCACC |
| CpbHLH025 | ACAAACAAGACAGATAGGGCAG | GAGAGTGGAATATCAGCAACCAG |
| CpbHLH030 | ACAAGCAAACCAACACTTCAG | CTCGTTCAGCACCTTGACC |
| CpbHLH055 | GTCAAGGTACTAAGCATGAGCA | TCTTCTTCCATCAGCTTTGTG |
| CpbHLH156 | AGCAACAGCGAGAATGGC | CAGCAGCTTCATCCTCTGG |
| CpbHLH158 | AGCTGGTTGCTGATATTCCAC | TCTTCCATCAGCTTTGCAACC |
| CpEE1a | ACTACGAGCAAGAGTTGGAAAC | TGGATGGCTGGAAGAGGAC |

图2 象草CpbHLH转录因子的保守基序分析a: CpbHLH基因家族的系统进化树Phylogenetic tree of the CpbHLH family; b: CpbHLH基因家族的基因结构Gene structure of the CpbHLH family; 绿色框、黑色线条分别代表外显子和内含子The green boxes, blacklines represent the exon and intron, respectively; c: CpbHLH基因家族的保守基序Conserved motifs of the CpbHLH family.
Fig.2 The conserved motifs analysis of C. purpureus bHLH transcription factors

图3 象草与水稻bHLH转录因子的系统进化分析分枝上的数字表示自展值Numbers on major branches indicate bootstrap estimates for 1000 replicate analyses. Cp: 象草C. purpureus; Os: 水稻O. sativa; A~U: 不同亚类Different subgroups.
Fig.3 Phylogenetic analysis of bHLH transcription factors in C. purpureus and O. sativa
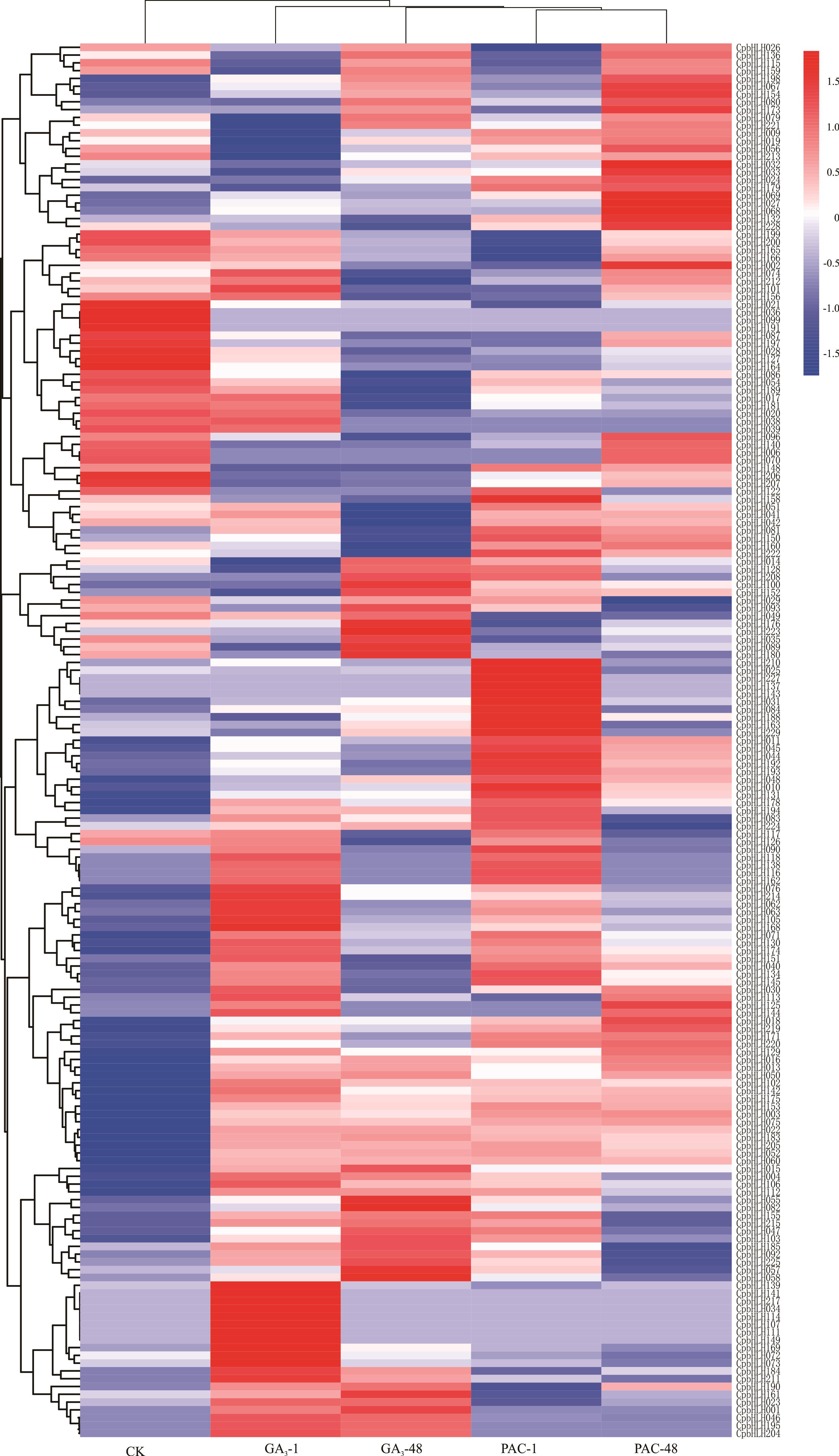
图4 象草CpbHLH基因在GA3和PAC处理1和48 h之后的表达分析红-白-蓝3色代表基因表达的强度,红色越亮代表表达量越高,蓝色越亮代表表达量越低,白色代表表达量为0 The three colors of red-white-blue represent the intensity of gene expression, the brighter the red, the higher the expression, the brighter the blue, the lower the expression, and the white represents the expression is 0. CK: 对照Control; GA3-1: GA3处理1 h GA3 treatment for 1 hour; GA3-48: GA3处理48 h GA3 treatment for 48 hour; PAC-1: PAC处理1 h PAC treatment for 1 hour; PAC-48: PAC处理48 h PAC treatment for 48 hour; 下同The same below.
Fig.4 Expression analysis of bHLH gene in C. purpureus after GA3 and PAC treatments for 1 and 48 hour
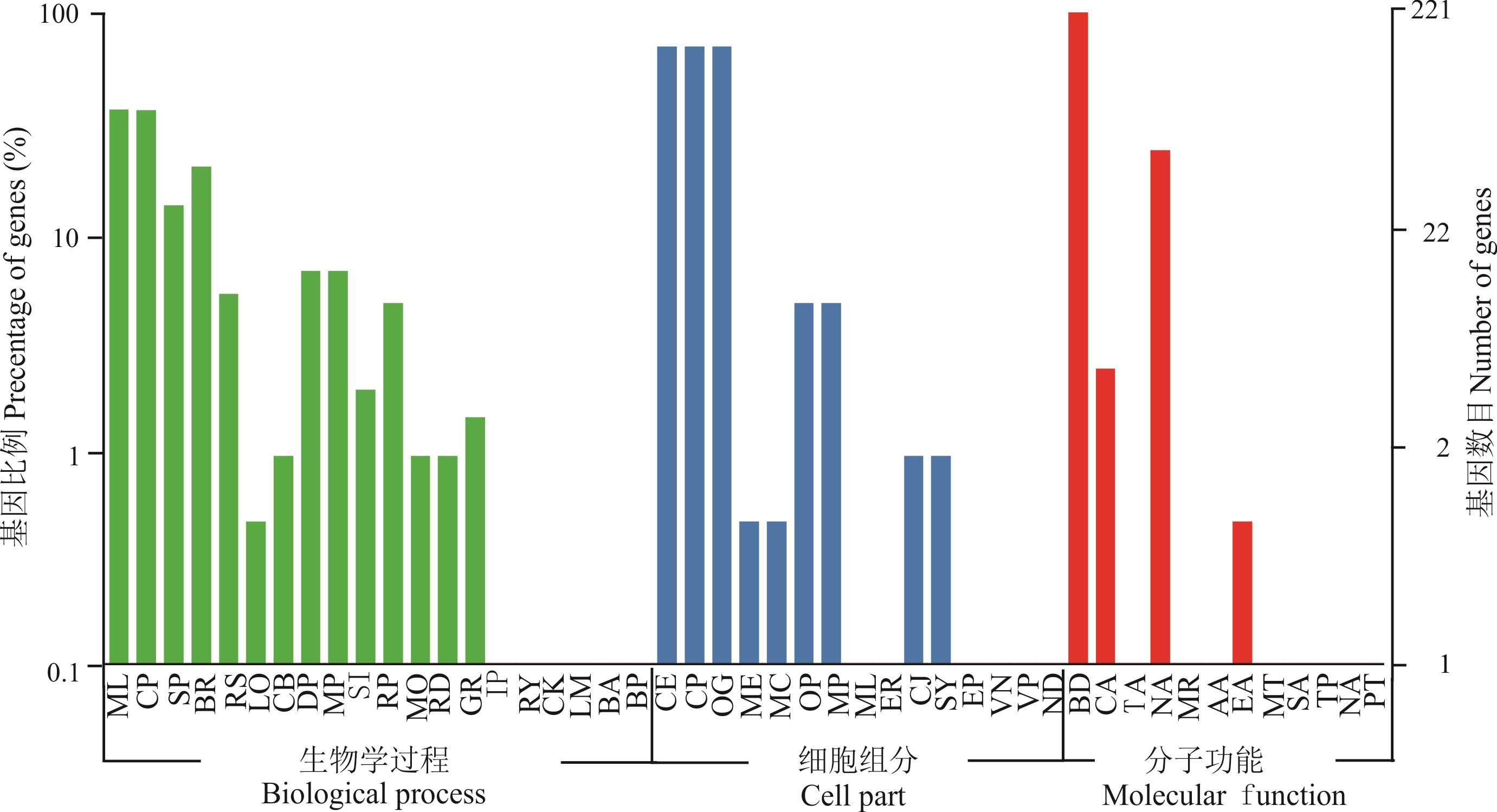
图6 象草bHLH基因的GO分类ML: 代谢过程Metabolic process; CP: 细胞过程Cellular process; SP: 单一有机体过程Single-organism process; BR: 生物调节Biological regulation; RS: 对刺激的响应Response to stimulus; LO: 定位Localization; CB: 细胞成分组织或生物组分Cellular component organization or biogenesis; DP: 发育过程 Developmental process; MP: 多细胞有机体过程Multicellular organismal process; SI: 信号Signaling; RP: 生殖过程Reproductive process; MO: 多有机体过程Multi-organism process; RD: 繁殖Reproduction; GR: 生长Growth; IP: 免疫系统过程Immune system process; RY: 节律过程Rhythmic process; CK: 细胞凋零Cell killing; LM: 运动Locomotion; BA: 生物附着Biological adhesion; BP: 生物相 Biological phase; CE: 细胞Cell; CP: 细胞组分Cell part; OG: 细胞器Organelle; ME: 细胞膜Membrane; MC: 膜部分Membrane part; OP: 器官部分Organelle part; MP: 大分子复合物Macromolecular complex; ML: 膜封闭腔Membrane-enclosed lumen; ER: 细胞外区域Extracellular region; CJ: 细胞结Cell junction; SY: 共质体Symplast; EP: 细胞外区域部分Extracellular region part; VN: 病毒Virion; VP: 病毒部分 Virion part; ND: 核Nucleoid; BD: 结合Binding; CA: 催化活性Catalytic activity; TA: 运输器活性Transporter activity; NA: 核酸结合转录因子活性Nucleic acid binding transcription factor activity; MR: 结构分子活性Structural molecule activity; AA: 抗氧化活性Antioxidant activity; EA: 电子载流子活性Electron carrier activity; MT: 分子传感器活性Molecular transducer activity; SA: 信号转导活性Signal transducer activity; TP: 转录因子活性,蛋白结合Transcription factor activity, protein binding; NA: 营养积累活性Nutrient reservoir activity; PT: 蛋白质标签Protein tag.
Fig.6 GO classification of CpbHLH genes
| 1 | Jiao W J. Study on extraction, purification, identification and properties of anthocyanins from purple elephant grass. Fuzhou: Fujian Agriculture and Forestry University, 2015. |
| 焦文静. 紫象草花色苷的提取纯化、结构分析及特性研究. 福州: 福建农林大学, 2015. | |
| 2 | Daud Z, Hatta M Z M, Kassim A S M, et al. Analysis of napier grass (Pennisetum purpureum) as a potential alternative fibre in paper industry. Material Research Innovations, 2014, 18(6): 18-20. |
| 3 | Huang Q F. The effect of additives on the quality of hybrid Pennisetum purpureum silage. Fuzhou: Fujian Agriculture and Forestry University, 2012. |
| 黄其芬. 不同添加剂对杂交象草青贮品质的影响. 福州: 福建农林大学, 2012. | |
| 4 | Wang Q B. Selection and breeding of hardy plant of Pennisetum purpureum schumach by bud radiation. Guizhou Agricultural Sciences, 2003, 31(6): 17-18. |
| 王庆斌. 象草辐照芽变选育耐寒植株的初步研究. 贵州农业科学, 2003, 31(6): 17-18. | |
| 5 | Zhang X, Gu H R, Ding C L, et al. Variation and cluster analysis of yield and biological characters of Pennisetum purpureum. Chinese Journal of Grassland, 2009, 31(1): 58-63. |
| 张霞, 顾洪如, 丁成龙, 等. 象草产量及生物学性状的品系间差异及聚类分析. 中国草地学报, 2009, 31(1): 58-63. | |
| 6 | Yi X F, Lai Z Q, Cai X Y, et al. High-yield cultivation, development and utilization of Pennisetum purpureum schumab cv. Red. Pruataculture & Animal Husbandry, 2012(1): 22-23. |
| 易显凤, 赖志强, 蔡小艳, 等. 紫色象草的高产栽培与开发利用. 草业与畜牧, 2012(1): 22-23. | |
| 7 | Alemayehu N, Abel T, Alok K, et al. Opportunities for napier grass (Pennisetum purpureum) improvement using molecular genetics. Agronomy, 2017, 7(2): 28. |
| 8 | Muktar M S, Teshome A, Hanson J, et al. Genotyping by sequencing provides new insights into the diversity of napier grass (Cenchrus purpureus) and reveals variation in genome-wide LD patterns between collections. Scientific Reports, 2019, 9(1): 1-15. |
| 9 | Wen X N, Jian Y Z, Xie X M. The comprehensive exploitation and utilization of Pennisetum purpureum. Pratacultural Science, 2005, 26(9): 108-112. |
| 温晓娜, 简有志, 解新明. 象草资源的综合开发利用. 草业科学, 2005, 26(9): 108-112. | |
| 10 | Ding D Y, Chen W D, Chen J X, et al. Comparative test on forage grass cultivars of Pennisetum. Guangdong Agricultural Sciences, 2019, 46(8): 8-13. |
| 丁迪云, 陈卫东, 陈杰雄, 等. 狼尾草属牧草品种比较试验. 广东农业科学, 2019, 46(8): 8-13. | |
| 11 | Hui Y K, Hu M L, Guo J Y, et al. The cloning and functional analysis of the transcription factor bHLH in purple-fleshed sweet potato. Journal of South China Normal University (Natural Science Edition), 2020, 52(4): 63-70. |
| 惠亚可, 胡敏伦, 郭晋雅, 等. 紫心甘薯转录因子bHLH的基因克隆及功能研究. 华南师范大学学报(自然科学版), 2020, 52(4): 63-70. | |
| 12 | Feng L, Shi Y B, Wang G B, et al. Bioinformatics and expression analysis of transcription factors of ginkgo bHLH family. Jiangsu Journal of Agricultural Science, 2019, 35(2): 400-411. |
| 冯磊, 石元豹, 汪贵斌, 等. 银杏bHLH家族转录因子生物信息学及表达分析. 江苏农业学报, 2019, 35(2): 400-411. | |
| 13 | Meng F X, Yang Y J, Duan Y J, et al. Identification and bioinformatics analysis of the bHLH transcription factor family in Citrus sinensis. Journal of Southwest Forestry University (Natural Science), 2020, 40(5): 73-86. |
| 孟富宣, 杨玉皎, 段元杰, 等. 甜橙bHLH转录因子家族的鉴定及生物信息学分析. 西南林业大学学报(自然科学), 2020, 40(5): 73-86. | |
| 14 | Zhang T, Lv W, Zhang H, et al. Genome-wide analysis of the basic helix-loop-helix (bHLH) transcription factor family in maize. BMC Plant Biology, 2018, 18(1): 235. |
| 15 | Mark E M, Cornelis M. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Molecular and Cellular Biology, 2000, 20(2): 429-440. |
| 16 | Chen H L, Hu L L, Wang L X, et al. Genome-wide identification and bioinformatics analysis of bHLH transcription factor family in mung bean (Vigna radiata L.). Journal of Plant Genetic Resources, 2017, 18(6): 1159-1167. |
| 陈红霖, 胡亮亮, 王丽侠, 等. 绿豆bHLH转录因子家族的鉴定与生物信息学分析. 植物遗传资源学报, 2017, 18(6): 1159-1167. | |
| 17 | Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell: Specific accumulation of flavonoids in Arabidopsis thaliana. Plant Journal, 2006, 46(5): 768-779. |
| 18 | Sun H, Fan H J, Ling H Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics, 2015, 16(1): 1-9. |
| 19 | Zhang X. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Beijing: Peking Union Medical College, 2015. |
| 张鑫. 基于丹参基因组和转录组发掘丹参酮生物合成相关的bHLH转录因子. 北京: 北京协和医学院, 2015. | |
| 20 | Komatsu K S, Maekawa M, Ujiie S, et al. LAX and SPA: Major regulators of shoot branching in rice. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(20): 11765-11770. |
| 21 | Gou J Y, Felippes F F, Liu C J, et al. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell, 2011, 23(4): 1512-1522. |
| 22 | Spelt C, Quattrocchio F, Mol J N, et al. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell, 2000, 12(9): 1619-1632. |
| 23 | Zhu T. Identification, phylogeny and expression profiling of bHLH transcription factor family in barley. Yangling: Northwest A&F University, 2019. |
| 朱婷. 大麦bHLH转录因子家族的鉴定及系统进化和表达分析. 杨凌: 西北农林科技大学, 2019. | |
| 24 | Heisler M G B, Atkinson A, Bylstra Y H, et al. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development, 2001, 128(7): 1089-1098. |
| 25 | He K P, Wu C. The effects of bHLH transcription factors on plant morphogenesis. Journal of Anhui Agricultural Science, 2010, 38(35): 19957-19959. |
| 何开平, 吴楚. bHLH转录因子对植物形态发生的影响. 安徽农业科学, 2010, 38(35): 19957-19959. | |
| 26 | Bailey P C, Martin C, Toledo-Ortiz G, et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell, 2003, 15(11): 2497-2501. |
| 27 | Li X X, Duan X P, Jiang H X, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiology, 2006, 141(4): 1167-1184. |
| 28 | Lorenzo C, Anahit G, Irma R, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology, 2010, 153(3): 1398-1412. |
| 29 | Li J. Study on agronomic traits and transcriptomics of exogenous gibberellin-induced internode elongation in Cenchrus purpureus. Lanzhou: Lanzhou University, 2020. |
| 李洁. 外源赤霉素诱导象草节间伸长的农艺性状和转录组学研究. 兰州: 兰州大学, 2020. | |
| 30 | Yan Q, Wu F, Xu P, et al. The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Molecular Ecology Resources, 2021, 21(2): 526-542. |
| 31 | Wu J Z, Qian C, Liu Z W, et al. De novo transcriptomic analysis for lignin synthesis in Cenchrus purpureus using RNA-seq. Acta Prataculturae Sinica, 2019, 28(1): 150-161. |
| 吴娟子, 钱晨, 刘智微, 等. 基于转录组测序分析象草木质素合成的研究. 草业学报, 2019, 28(1): 150-161. | |
| 32 | Zhang J Y, Wang Y R, Nan Z B. Relative and absolute quantification-expression analysis of CsSAMDC gene as a case. China Biotechnology, 2009, 29(8): 86-91. |
| 张吉宇, 王彦荣, 南志标. 相对定量和绝对定量-以CsSAMDC基因表达分析为例. 中国生物工程杂志, 2009, 29(8): 86-91. | |
| 33 | Yang M T, Zhang C, Wang Z P, et al. Cloning and functional analysis of ZmbHLH161 gene in maize. Acta Agronomica Sinica, 2020, 46(12): 194-202. |
| 杨梦婷, 张春, 王作平, 等. 玉米ZmbHLH161基因的克隆及功能研究. 作物学报, 2020, 46(12): 194-202. | |
| 34 | Pei L Q. Identification and preliminary analysis of bHLH gene family in sweet potato. Xuzhou: Jiangsu Normal University, 2017. |
| 裴苓荃. 甘薯bHLH基因家族的鉴定与初步分析. 徐州: 江苏师范大学, 2017. | |
| 35 | Gabriela T, Enamul H, Peter H Q. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell, 2003, 15(8): 1749-1770. |
| 36 | Xin N, Guan Y, Chen S, et al. Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genomics, 2017, 18(1): 619. |
| 37 | Zhang S H, Qu L H. The evolution of genome and genes in introns. Acta Scientiarum Naturalium Universitatis Sunyatseni (Natural Science Edition), 1999, 38(1): 51-55. |
| 张尚宏, 屈良鹄. 基因组的进化与内含子中的基因的进化. 中山大学学报(自然科学版), 1999, 38(1): 51-55. | |
| 38 | Xu G, Guo C, Shan H, et al. Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences, 2012, 109(4): 1187-1192. |
| 39 | Yang J H. Identification and expression of apple basic helix-loop-helix transcripton factory family. Yangling: Northwest A&F University, 2017 |
| 杨金华. 苹果bHLH转录因子家族的鉴定及表达分析. 杨凌: 西北农林科技大学, 2017. | |
| 40 | Pires N, Dolan L. Early evolution of bHLH proteins in plants. Plant Signaling & Behavior, 2010, 5(7): 911-912. |
| 41 | Song X M, Huang Z N, Duan W K, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Molecular Genetics Genomics, 2014, 289(1): 77-91. |
| 42 | Wang J, Hu Z, Zhao T, et al. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics, 2015, 16(1): 39. |
| 43 | Xu X R, Yang K B, Wang S N, et al. Identification of bHLH transcription factors in moso bamboo (Phyllostachys edulis) and their expression analysis under drought and salt stress. Plant Science Journal, 2019, 37(5): 610-620. |
| 徐秀荣, 杨克彬, 王思宁, 等. 毛竹bHLH转录因子的鉴定及其在干旱和盐胁迫条件下的表达分析. 植物科学学报, 2019, 37(5): 610-620. | |
| 44 | He J, Gu X R, Wei C H, et al. Identification and expression analysis under abiotic stresses of the bHLH transcription factor gene family in watermelon. Acta Horticulturae Sinica, 2016, 43(2): 281-294. |
| 何洁, 顾秀容, 魏春华, 等. 西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析. 园艺学报, 2016, 43(2): 281-294. | |
| 45 | Nuno P, Liam D. Origin and diversification of basic-helix-loop-helix proteins in plants. Molecular Biology and Evolution, 2010, 27(4): 862-874. |
| 46 | Chen H C, Cheng W H, Hong C Y, et al. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice, 2018, 11(1): 1-17. |
| 47 | Wen J, Li Y, Qi T, et al. The C-terminal domains of Arabidopsis GL3/EGL3/TT8 interact with JAZ proteins and mediate dimeric interactions. Plant Signaling & Behavior, 2018, 13(1): e1422460. |
| [1] | 魏娜, 李艳鹏, 马艺桐, 刘文献. 全基因组水平紫花苜蓿TCP基因家族的鉴定及其在干旱胁迫下表达模式分析[J]. 草业学报, 2022, 31(1): 118-130. |
| [2] | 尹祥, 王咏琪, 李鑫琴, 田静, 王晓亚, 张建国. 不同水分吸附材料对象草青贮发酵品质及好氧稳定性的影响[J]. 草业学报, 2021, 30(7): 133-138. |
| [3] | 罗维, 舒健虹, 刘晓霞, 王子苑, 牟琼, 王小利, 吴佳海. 高羊茅FaRVE8基因的克隆、亚细胞定位及表达分析[J]. 草业学报, 2020, 29(7): 60-69. |
| [4] | 杨婷, 张建平, 刘自刚, 齐燕妮, 李闻娟, 谢亚萍. 胡麻异质型ACCase亚基基因的克隆与表达分析[J]. 草业学报, 2020, 29(4): 111-120. |
| [5] | 夏曾润, 王文颖, 刘亚琪, 王锁民. 罗布麻K+通道编码基因AvAKT1的克隆与表达分析[J]. 草业学报, 2019, 28(8): 180-189. |
| [6] | 滕珂, 张蕊, 檀鹏辉, 岳跃森, 范希峰, 武菊英. 日本结缕草ZjERF1的克隆、转录激活活性、亚细胞定位及表达分析[J]. 草业学报, 2019, 28(6): 56-65. |
| [7] | 陆姗姗, 洪园淑, 刘萍. 苦豆子赖氨酸脱羧酶基因启动子在拟南芥中的表达分析[J]. 草业学报, 2019, 28(11): 159-167. |
| [8] | 李闻娟, 齐燕妮, 王利民, 党照, 赵利, 赵玮, 谢亚萍, 王斌, 张建平, 李淑洁. 不同胡麻品种TAG合成途径关键基因表达与含油量、脂肪酸组分的相关性分析[J]. 草业学报, 2019, 28(1): 138-149. |
| [9] | 吴娟子, 钱晨, 刘智微, 潘玉梅, 钟小仙. 基于转录组测序分析象草木质素合成的研究[J]. 草业学报, 2019, 28(1): 150-161. |
| [10] | 贾俊婷, 赵品苍, 刘祝江, 袁光孝, 杨伟光, 刘书, 陈双燕, 李晓霞, 刘公社. 羊草LcMADS12基因的克隆及原核表达分析[J]. 草业学报, 2018, 27(7): 64-72. |
| [11] | 吴征敏, 王志敬, 吴浩浩, 李政, 李文威, 庄桂锋, 尹福泉, 赵志辉. 象草与皇竹草组合效应的研究[J]. 草业学报, 2018, 27(2): 135-145. |
| [12] | 武炳超, 张欢, 童磊, 杜昭昌, 胡家菱, 陈燚, 张新全, 刘伟, 黄琳凯. 象草不同辐射剂量诱变系表型及遗传变异研究[J]. 草业学报, 2018, 27(11): 77-86. |
| [13] | 董笛, 滕珂, 于安东, 檀鹏辉, 梁小红, 韩烈保. 沟叶结缕草八氢番茄红素基因ZmPSY的克隆、亚细胞定位及表达分析[J]. 草业学报, 2017, 26(11): 69-76. |
| [14] | 邵麟惠, 郑兴卫, 李聪. 蒺藜苜蓿E3泛素连接酶U-box基因克隆及表达分[J]. 草业学报, 2016, 25(7): 62-72. |
| [15] | 张胤冰, 孙鑫博, 樊波, 韩烈保, 张雪, 袁建波, 许立新. 结缕草ZjNAC基因的克隆与表达分析[J]. 草业学报, 2016, 25(4): 239-245. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||