

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (4): 171-185.DOI: 10.11686/cyxb2023192
• 研究论文 • 上一篇
刘保财1,2,3( ), 胡学博2, 张武君1,3, 赵云青1,3, 黄颖桢1,3, 陈菁瑛1,3(
), 胡学博2, 张武君1,3, 赵云青1,3, 黄颖桢1,3, 陈菁瑛1,3( )
)
收稿日期:2023-06-09
修回日期:2023-08-04
出版日期:2024-04-20
发布日期:2024-01-15
通讯作者:
陈菁瑛
作者简介:E-mail: cjy6601@163.com基金资助:
Bao-cai LIU1,2,3( ), Xue-bo HU2, Wu-jun ZHANG1,3, Yun-qing ZHAO1,3, Ying-zhen HUANG1,3, Jing-ying CHEN1,3(
), Xue-bo HU2, Wu-jun ZHANG1,3, Yun-qing ZHAO1,3, Ying-zhen HUANG1,3, Jing-ying CHEN1,3( )
)
Received:2023-06-09
Revised:2023-08-04
Online:2024-04-20
Published:2024-01-15
Contact:
Jing-ying CHEN
摘要:
石仙桃和细叶石仙桃均为珍稀濒危的附生兰,两者叶绿体基因组特征及其系统发育报道较少,本研究旨在揭示石仙桃和细叶石仙桃叶绿体基因组特征及其系统发育。2022年5月取驯化栽培的石仙桃和细叶石仙桃幼嫩叶片,基于Illumina测序平台分别获得石仙桃和细叶石仙桃叶绿体序列,用GetOrganelle组装、CPGAVAS2注释,利用生物信息学方法开展重复序列、密码子偏好性、四分体边界等叶绿体特征分析,并从NCBI上下载相关的序列开展基因组比较与系统发育分析。石仙桃叶绿体基因组大小为159122 bp(GC含量为37.41%),细叶石仙桃叶绿体基因组大小为158798 bp(GC含量为37.47%),两者均包括大单拷贝区(LSC)、小单拷贝区(SSC)和反向重复序列(IRa/IRb)。石仙桃与细叶石仙桃均注释了113个基因,其中编码蛋白质的基因79个,石仙桃中编码79个蛋白质基因使用22968个密码子,编码亮氨酸的UUA相对同义密码子使用频次(RSCU)达到1.90;细叶石仙桃中编码79个蛋白质的基因使用22923个密码子,编码亮氨酸UUA的RSCU达到1.91。石仙桃叶绿体基因组中共有44个简单序列重复(SSR)、47个散在重复、26个串联重复;细叶石仙桃有32个SSR、25个散在重复、30个串联重复。石仙桃属6个物种的四分体边界及共线性无大片段变化,但存在核苷酸多态性,trnK-UUU、trnQ-UUG、rpl16等基因可作为物种的分子标记候选基因。系统分析明确了石仙桃和细叶石仙桃等石仙桃属物种与蜂腰兰和流苏贝母亲缘关系较近,并同处于附生兰的分支中,叶绿体基因组大片段倒置等可能是兰科适应生活习性改变的原因之一。本研究明确了石仙桃和细叶石仙桃叶绿体基因组特征,探讨了其系统发育及兰科生活习性与叶绿体基因组之间的关系,为进一步分类和分子标记开发等相关研究奠定了基础。
刘保财, 胡学博, 张武君, 赵云青, 黄颖桢, 陈菁瑛. 石仙桃与细叶石仙桃叶绿体基因组解析及其系统发育[J]. 草业学报, 2024, 33(4): 171-185.
Bao-cai LIU, Xue-bo HU, Wu-jun ZHANG, Yun-qing ZHAO, Ying-zhen HUANG, Jing-ying CHEN. Chloroplast genome assembly and phylogenetic analysis of Pholidota chinensis and Pholidota cantonensis[J]. Acta Prataculturae Sinica, 2024, 33(4): 171-185.
| 基因Genes | 列表List | 数量No. |
|---|---|---|
| 自我复制相关基因Genes for self-replication | ||
| 核糖体RNA基因rRNA genes | rrn16, rrn23, rrn4.5, rrn5 | 4 |
| 转运RNA基因tRNA genes | trnA-UGC*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU, trnI-GAU*, trnK-UUU*, trnL-CAA, trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | 30 |
| 核糖体小亚基基因Small subunit of ribosome genes | rps11, rps12**, rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | 12 |
| 核糖体大亚基基因Large subunit of ribosome genes | rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | 9 |
| RNA 聚合酶亚基基因RNA polymerase subunit genes | rpoA, rpoB, rpoC1*, rpoC2 | 4 |
| 光合作用相关基因Genes for photosynthesis | ||
| NADH 脱氢酶基因NADH dehydrogenase genes | ndhA*, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 11 |
| ATP 合成酶基因ATP synthase genes | atpA, atpB, atpE, atpF*, atpH, atpI | 6 |
| 光合系统Ⅰ基因Photosystem I genes | psaA, psaB, psaC, psaI, psaJ | 5 |
| 光合系统Ⅱ基因Photosystem Ⅱ genes | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 细胞色素b/f复合体基因Cytochrome b/f complex genes | petA, petB*, petD*, petG, petL, petN | 6 |
| RubisCO大亚基基因RubisCO large subunit gene | rbcL | 1 |
| 其他基因Other genes | ||
| 成熟酶K基因Maturase K gene | matK | 1 |
| 蛋白酶基因Protease gene | clpP** | 1 |
| 囊膜蛋白基因Envelop membrane protein gene | cemA | 1 |
| C 型细胞色素合成基因C-type cytochrome synthesis gene | ccsA | 1 |
| 翻译起始因子基因Translational initiation factor gene | infA | 1 |
乙酰辅酶A羧化酶亚基基因 Acetyl-CoA carboxylase beta subunit gene | accD | 1 |
| 未知功能基因Unknown function genes | ||
假定叶绿体阅读框基因 Hypothetical chloroplast reading frames genes | ycf1, ycf2, ycf3**, ycf4 | 4 |
表1 石仙桃和细叶石仙桃叶绿体基因组功能注释与分类
Table 1 Gene functional annotation and classification of chloroplast genome in P. chinensis and P. cantonensis
| 基因Genes | 列表List | 数量No. |
|---|---|---|
| 自我复制相关基因Genes for self-replication | ||
| 核糖体RNA基因rRNA genes | rrn16, rrn23, rrn4.5, rrn5 | 4 |
| 转运RNA基因tRNA genes | trnA-UGC*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU, trnI-GAU*, trnK-UUU*, trnL-CAA, trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | 30 |
| 核糖体小亚基基因Small subunit of ribosome genes | rps11, rps12**, rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | 12 |
| 核糖体大亚基基因Large subunit of ribosome genes | rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | 9 |
| RNA 聚合酶亚基基因RNA polymerase subunit genes | rpoA, rpoB, rpoC1*, rpoC2 | 4 |
| 光合作用相关基因Genes for photosynthesis | ||
| NADH 脱氢酶基因NADH dehydrogenase genes | ndhA*, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 11 |
| ATP 合成酶基因ATP synthase genes | atpA, atpB, atpE, atpF*, atpH, atpI | 6 |
| 光合系统Ⅰ基因Photosystem I genes | psaA, psaB, psaC, psaI, psaJ | 5 |
| 光合系统Ⅱ基因Photosystem Ⅱ genes | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 细胞色素b/f复合体基因Cytochrome b/f complex genes | petA, petB*, petD*, petG, petL, petN | 6 |
| RubisCO大亚基基因RubisCO large subunit gene | rbcL | 1 |
| 其他基因Other genes | ||
| 成熟酶K基因Maturase K gene | matK | 1 |
| 蛋白酶基因Protease gene | clpP** | 1 |
| 囊膜蛋白基因Envelop membrane protein gene | cemA | 1 |
| C 型细胞色素合成基因C-type cytochrome synthesis gene | ccsA | 1 |
| 翻译起始因子基因Translational initiation factor gene | infA | 1 |
乙酰辅酶A羧化酶亚基基因 Acetyl-CoA carboxylase beta subunit gene | accD | 1 |
| 未知功能基因Unknown function genes | ||
假定叶绿体阅读框基因 Hypothetical chloroplast reading frames genes | ycf1, ycf2, ycf3**, ycf4 | 4 |

图2 石仙桃和细叶石仙桃叶绿体基因组相对同义密码子使用频次(RSCU)A: 丙氨酸Alanine (Ala); C: 半胱氨酸L-cysteine (Cys); D: 天冬氨酸Aspartic acid (Asp); E: 谷氨酸Glutamate (Glu); F: 苯丙氨酸Phenylalanine (Phe); G: 甘氨酸Glycine (Gly); H: 组氨酸Histidine (His); I: 异亮氨酸Isoleucine (Iso); K: 赖氨酸Lysine (Lys); L: 亮氨酸Leucine (Leu); M: 蛋氨酸Methionine (Met); N: 天冬酰胺Asparagine (Asp); P: 脯氨酸Proline (Pro); Q: 谷氨酰胺Glutamin (Glu); R: 精氨酸Arginine (Arg); S: 丝氨酸Serine (Ser); U: 苏氨酸Threonine (Thr); V: 缬氨酸Valine (Val); W: 色氨酸Tryptophan (Try); Y: 酪氨酸Tyrosine (Tyr); Z: 终止密码子The stop codon.
Fig. 2 Relative synonymous codon usage (RSCU) of P. chinensis and P. cantonensis
物种 Species | RL (bp) | MD | 起始位置Starting position (bp) | 物种 Species | RL (bp) | MD | 起始位置Starting position (bp) | ||
|---|---|---|---|---|---|---|---|---|---|
| 重复1 Repeat 1 | 重复2 Repeat 2 | 重复1 Repeat 1 | 重复2 Repeat 2 | ||||||
| PCH | 26699 | P | 86910 | 132423 | PCH | 31 | P | 94747 | 151278 |
| PCH | 59 | P | 13 | 13 | PCH | 31 | P | 99062 | 99062 |
| PCH | 50 | P | 125188 | 125188 | PCH | 31 | F | 99062 | 146939 |
| PCH | 46 | P | 30245 | 30245 | PCH | 31 | P | 146939 | 146939 |
| PCH | 49 | F | 92277 | 92298 | PCH | 31 | F | 151257 | 151281 |
| PCH | 49 | P | 92277 | 153685 | PCH | 30 | F | 10149 | 37527 |
| PCH | 49 | P | 92298 | 153706 | PCH | 30 | F | 29317 | 69320 |
| PCH | 49 | F | 153685 | 153706 | PCH | 30 | F | 39780 | 42004 |
| PCH | 47 | P | 64797 | 64797 | PCH | 30 | F | 44756 | 102157 |
| PCH | 35 | F | 94706 | 94748 | PCH | 30 | P | 44756 | 143845 |
| PCH | 35 | P | 94706 | 151249 | PCH | 30 | P | 60483 | 60521 |
| PCH | 35 | P | 94748 | 151291 | PCA | 26514 | P | 87008 | 132284 |
| PCH | 35 | F | 151249 | 151291 | PCA | 46 | P | 30029 | 30029 |
| PCH | 34 | P | 5271 | 5271 | PCA | 53 | P | 64931 | 64931 |
| PCH | 39 | F | 44744 | 102145 | PCA | 45 | P | 0 | 0 |
| PCH | 39 | P | 44744 | 143848 | PCA | 34 | P | 5233 | 5233 |
| PCH | 30 | P | 37602 | 37602 | PCA | 39 | F | 44829 | 102053 |
| PCH | 30 | P | 118249 | 118249 | PCA | 39 | P | 44829 | 143714 |
| PCH | 37 | P | 127707 | 127707 | PCA | 38 | P | 125153 | 125153 |
| PCH | 37 | P | 129089 | 129089 | PCA | 30 | P | 118177 | 118177 |
| PCH | 30 | P | 8563 | 46180 | PCA | 37 | P | 128923 | 128923 |
| PCH | 31 | F | 94702 | 94720 | PCA | 34 | P | 3432 | 3432 |
| PCH | 31 | P | 94702 | 151281 | PCA | 30 | P | 8383 | 46270 |
| PCH | 31 | P | 94720 | 151299 | PCA | 33 | P | 12697 | 12697 |
| PCH | 31 | F | 151281 | 151299 | PCA | 30 | P | 6168 | 6168 |
| PCH | 33 | P | 12873 | 12873 | PCA | 32 | P | 36907 | 46270 |
| PCH | 30 | P | 6196 | 6196 | PCA | 31 | P | 98970 | 98970 |
| PCH | 30 | P | 43273 | 76389 | PCA | 31 | F | 98970 | 146805 |
| PCH | 30 | P | 47739 | 47739 | PCA | 31 | P | 146805 | 146805 |
| PCH | 30 | F | 92277 | 92319 | PCA | 30 | F | 9970 | 37740 |
| PCH | 30 | P | 92277 | 153683 | PCA | 30 | F | 29081 | 69448 |
| PCH | 30 | P | 92319 | 153725 | PCA | 30 | F | 39874 | 42098 |
| PCH | 30 | F | 153685 | 153727 | PCA | 30 | F | 43395 | 116545 |
| PCH | 32 | P | 36760 | 46180 | PCA | 30 | F | 44841 | 102065 |
| PCH | 31 | F | 94723 | 94747 | PCA | 30 | P | 44841 | 143711 |
| PCH | 31 | P | 94723 | 151254 | PCA | 30 | P | 60617 | 60655 |
表2 石仙桃和细叶石仙桃叶绿体基因组中的重复序列
Table 2 Repeat sequences of chloroplast genome in P. chinensis and P. cantonensis
物种 Species | RL (bp) | MD | 起始位置Starting position (bp) | 物种 Species | RL (bp) | MD | 起始位置Starting position (bp) | ||
|---|---|---|---|---|---|---|---|---|---|
| 重复1 Repeat 1 | 重复2 Repeat 2 | 重复1 Repeat 1 | 重复2 Repeat 2 | ||||||
| PCH | 26699 | P | 86910 | 132423 | PCH | 31 | P | 94747 | 151278 |
| PCH | 59 | P | 13 | 13 | PCH | 31 | P | 99062 | 99062 |
| PCH | 50 | P | 125188 | 125188 | PCH | 31 | F | 99062 | 146939 |
| PCH | 46 | P | 30245 | 30245 | PCH | 31 | P | 146939 | 146939 |
| PCH | 49 | F | 92277 | 92298 | PCH | 31 | F | 151257 | 151281 |
| PCH | 49 | P | 92277 | 153685 | PCH | 30 | F | 10149 | 37527 |
| PCH | 49 | P | 92298 | 153706 | PCH | 30 | F | 29317 | 69320 |
| PCH | 49 | F | 153685 | 153706 | PCH | 30 | F | 39780 | 42004 |
| PCH | 47 | P | 64797 | 64797 | PCH | 30 | F | 44756 | 102157 |
| PCH | 35 | F | 94706 | 94748 | PCH | 30 | P | 44756 | 143845 |
| PCH | 35 | P | 94706 | 151249 | PCH | 30 | P | 60483 | 60521 |
| PCH | 35 | P | 94748 | 151291 | PCA | 26514 | P | 87008 | 132284 |
| PCH | 35 | F | 151249 | 151291 | PCA | 46 | P | 30029 | 30029 |
| PCH | 34 | P | 5271 | 5271 | PCA | 53 | P | 64931 | 64931 |
| PCH | 39 | F | 44744 | 102145 | PCA | 45 | P | 0 | 0 |
| PCH | 39 | P | 44744 | 143848 | PCA | 34 | P | 5233 | 5233 |
| PCH | 30 | P | 37602 | 37602 | PCA | 39 | F | 44829 | 102053 |
| PCH | 30 | P | 118249 | 118249 | PCA | 39 | P | 44829 | 143714 |
| PCH | 37 | P | 127707 | 127707 | PCA | 38 | P | 125153 | 125153 |
| PCH | 37 | P | 129089 | 129089 | PCA | 30 | P | 118177 | 118177 |
| PCH | 30 | P | 8563 | 46180 | PCA | 37 | P | 128923 | 128923 |
| PCH | 31 | F | 94702 | 94720 | PCA | 34 | P | 3432 | 3432 |
| PCH | 31 | P | 94702 | 151281 | PCA | 30 | P | 8383 | 46270 |
| PCH | 31 | P | 94720 | 151299 | PCA | 33 | P | 12697 | 12697 |
| PCH | 31 | F | 151281 | 151299 | PCA | 30 | P | 6168 | 6168 |
| PCH | 33 | P | 12873 | 12873 | PCA | 32 | P | 36907 | 46270 |
| PCH | 30 | P | 6196 | 6196 | PCA | 31 | P | 98970 | 98970 |
| PCH | 30 | P | 43273 | 76389 | PCA | 31 | F | 98970 | 146805 |
| PCH | 30 | P | 47739 | 47739 | PCA | 31 | P | 146805 | 146805 |
| PCH | 30 | F | 92277 | 92319 | PCA | 30 | F | 9970 | 37740 |
| PCH | 30 | P | 92277 | 153683 | PCA | 30 | F | 29081 | 69448 |
| PCH | 30 | P | 92319 | 153725 | PCA | 30 | F | 39874 | 42098 |
| PCH | 30 | F | 153685 | 153727 | PCA | 30 | F | 43395 | 116545 |
| PCH | 32 | P | 36760 | 46180 | PCA | 30 | F | 44841 | 102065 |
| PCH | 31 | F | 94723 | 94747 | PCA | 30 | P | 44841 | 143711 |
| PCH | 31 | P | 94723 | 151254 | PCA | 30 | P | 60617 | 60655 |
位置 Position (bp) | 片段 Fragment (bp) | 重复 Copy | 一致大小 Consensus size (bp) | 匹配比 Percent matches (%) | 插入删除 比率Percent indels (%) | 比例 Percent (%) | 熵 Entropy (0~2) | 一致性序列 Consistent sequence | |
|---|---|---|---|---|---|---|---|---|---|
| A | C | ||||||||
| 1634~1670 | 19 | 2.0 | 18 | 89 | 5 | 43 | 8 | 1.74 | ACGAATATCATGATATAT |
| 4809~4845 | 18 | 2.1 | 17 | 90 | 5 | 78 | 0 | 0.96 | AAAAAAAAGAAGAAAAT |
| 6480~6522 | 16 | 2.6 | 16 | 85 | 3 | 53 | 16 | 1.43 | TAACTAAATAATATAC |
| 14554~14578 | 12 | 2.1 | 12 | 100 | 0 | 24 | 24 | 1.80 | TTCTGTAACCAT |
| 15813~15837 | 12 | 2.1 | 12 | 100 | 0 | 48 | 0 | 1.46 | TTAATGATAAGA |
| 29285~29339 | 21 | 2.6 | 20 | 76 | 18 | 18 | 9 | 1.10 | TTTTTTTTTCTTATATTCTA |
| 29286~29359 | 22 | 3.4 | 21 | 78 | 14 | 18 | 8 | 1.18 | TTTTATTTCTTATATTCTATA |
| 51318~51342 | 9 | 2.8 | 9 | 100 | 0 | 60 | 8 | 1.57 | AAGAAATTC |
| 55676~55737 | 10 | 6.1 | 10 | 82 | 17 | 30 | 8 | 1.25 | TCTATTTATA |
| 55676~55739 | 22 | 3.0 | 20 | 81 | 18 | 31 | 7 | 1.25 | TCTATTTATATTATTTTATA |
| 55752~55783 | 16 | 2.0 | 16 | 93 | 0 | 9 | 21 | 1.17 | TCTTTATCTTTACTTT |
| 69771~69800 | 15 | 2.0 | 15 | 93 | 0 | 63 | 13 | 1.50 | AAGGAATAAACAAAC |
| 69807~69850 | 21 | 2.1 | 21 | 86 | 0 | 34 | 25 | 1.87 | TAAATCCAAACAACCTTTTCG |
| 79827~79866 | 20 | 2.0 | 20 | 100 | 0 | 45 | 0 | 1.51 | ATATGTAGATATGTATGAAA |
| 83372~83423 | 27 | 2.0 | 25 | 85 | 14 | 11 | 5 | 1.13 | TTCTTTTTTTTCTTTTTTTTTTGAA |
| 92258~92347 | 21 | 4.1 | 21 | 92 | 4 | 13 | 26 | 1.73 | TTTGTCCAAGTCACTTCTCTT |
| 94700~94787 | 9 | 10.1 | 9 | 78 | 7 | 32 | 4 | 1.77 | TATTGATGA |
| 94707~94783 | 18 | 3.9 | 18 | 81 | 18 | 33 | 5 | 1.79 | GATATTGATGATAGTGAC |
| 94724~94771 | 24 | 2.0 | 24 | 100 | 0 | 33 | 8 | 1.86 | CGATATTGATGATAGTGACGATAT |
| 94724~94784 | 24 | 2.5 | 24 | 89 | 5 | 32 | 6 | 1.82 | CGATATAGATGATAGTGACGATAT |
| 116164~116201 | 17 | 2.2 | 17 | 86 | 9 | 34 | 7 | 1.28 | ATATTATATATATTTCT |
| 116300~116345 | 15 | 3.1 | 15 | 87 | 9 | 19 | 13 | 1.23 | TTTATTCTTTACTAT |
| 130054~130080 | 12 | 2.2 | 12 | 100 | 0 | 14 | 11 | 1.08 | TTCTTTATTTAT |
| 151250~151326 | 18 | 3.9 | 18 | 81 | 18 | 33 | 27 | 1.79 | TCACTATCATCAATATCG |
| 151246~151333 | 9 | 10.1 | 9 | 75 | 7 | 36 | 26 | 1.77 | ATCATCAAT |
| 153686~153775 | 21 | 4.1 | 21 | 92 | 4 | 50 | 10 | 1.73 | AAAAAGAGAAGTGACTTGGAC |
表3 石仙桃叶绿体基因组中串联重复序列
Table 3 Tandem repeat sequences of chloroplast genome in P. chinensis
位置 Position (bp) | 片段 Fragment (bp) | 重复 Copy | 一致大小 Consensus size (bp) | 匹配比 Percent matches (%) | 插入删除 比率Percent indels (%) | 比例 Percent (%) | 熵 Entropy (0~2) | 一致性序列 Consistent sequence | |
|---|---|---|---|---|---|---|---|---|---|
| A | C | ||||||||
| 1634~1670 | 19 | 2.0 | 18 | 89 | 5 | 43 | 8 | 1.74 | ACGAATATCATGATATAT |
| 4809~4845 | 18 | 2.1 | 17 | 90 | 5 | 78 | 0 | 0.96 | AAAAAAAAGAAGAAAAT |
| 6480~6522 | 16 | 2.6 | 16 | 85 | 3 | 53 | 16 | 1.43 | TAACTAAATAATATAC |
| 14554~14578 | 12 | 2.1 | 12 | 100 | 0 | 24 | 24 | 1.80 | TTCTGTAACCAT |
| 15813~15837 | 12 | 2.1 | 12 | 100 | 0 | 48 | 0 | 1.46 | TTAATGATAAGA |
| 29285~29339 | 21 | 2.6 | 20 | 76 | 18 | 18 | 9 | 1.10 | TTTTTTTTTCTTATATTCTA |
| 29286~29359 | 22 | 3.4 | 21 | 78 | 14 | 18 | 8 | 1.18 | TTTTATTTCTTATATTCTATA |
| 51318~51342 | 9 | 2.8 | 9 | 100 | 0 | 60 | 8 | 1.57 | AAGAAATTC |
| 55676~55737 | 10 | 6.1 | 10 | 82 | 17 | 30 | 8 | 1.25 | TCTATTTATA |
| 55676~55739 | 22 | 3.0 | 20 | 81 | 18 | 31 | 7 | 1.25 | TCTATTTATATTATTTTATA |
| 55752~55783 | 16 | 2.0 | 16 | 93 | 0 | 9 | 21 | 1.17 | TCTTTATCTTTACTTT |
| 69771~69800 | 15 | 2.0 | 15 | 93 | 0 | 63 | 13 | 1.50 | AAGGAATAAACAAAC |
| 69807~69850 | 21 | 2.1 | 21 | 86 | 0 | 34 | 25 | 1.87 | TAAATCCAAACAACCTTTTCG |
| 79827~79866 | 20 | 2.0 | 20 | 100 | 0 | 45 | 0 | 1.51 | ATATGTAGATATGTATGAAA |
| 83372~83423 | 27 | 2.0 | 25 | 85 | 14 | 11 | 5 | 1.13 | TTCTTTTTTTTCTTTTTTTTTTGAA |
| 92258~92347 | 21 | 4.1 | 21 | 92 | 4 | 13 | 26 | 1.73 | TTTGTCCAAGTCACTTCTCTT |
| 94700~94787 | 9 | 10.1 | 9 | 78 | 7 | 32 | 4 | 1.77 | TATTGATGA |
| 94707~94783 | 18 | 3.9 | 18 | 81 | 18 | 33 | 5 | 1.79 | GATATTGATGATAGTGAC |
| 94724~94771 | 24 | 2.0 | 24 | 100 | 0 | 33 | 8 | 1.86 | CGATATTGATGATAGTGACGATAT |
| 94724~94784 | 24 | 2.5 | 24 | 89 | 5 | 32 | 6 | 1.82 | CGATATAGATGATAGTGACGATAT |
| 116164~116201 | 17 | 2.2 | 17 | 86 | 9 | 34 | 7 | 1.28 | ATATTATATATATTTCT |
| 116300~116345 | 15 | 3.1 | 15 | 87 | 9 | 19 | 13 | 1.23 | TTTATTCTTTACTAT |
| 130054~130080 | 12 | 2.2 | 12 | 100 | 0 | 14 | 11 | 1.08 | TTCTTTATTTAT |
| 151250~151326 | 18 | 3.9 | 18 | 81 | 18 | 33 | 27 | 1.79 | TCACTATCATCAATATCG |
| 151246~151333 | 9 | 10.1 | 9 | 75 | 7 | 36 | 26 | 1.77 | ATCATCAAT |
| 153686~153775 | 21 | 4.1 | 21 | 92 | 4 | 50 | 10 | 1.73 | AAAAAGAGAAGTGACTTGGAC |
位置 Position (bp) | 片段 Fragment (bp) | 重复Copy | 一致 大小 Consensus size (bp) | 匹配比率 Percent matches (%) | 插入删除比率Percent indels (%) | 比例 Percent (%) | 熵 Entropy (0~2) | 一致性序列 Consistent sequence | |
|---|---|---|---|---|---|---|---|---|---|
| A | C | ||||||||
| 3690~3724 | 17 | 2.1 | 17 | 88 | 0 | 8 | 11 | 1.31 | ATTTTCTTTTCCTTGTT |
| 4759~4802 | 18 | 2.4 | 18 | 85 | 7 | 75 | 2 | 1.03 | AAGAAGAAGAAGAAAAAA |
| 4766~4808 | 20 | 2.2 | 20 | 87 | 8 | 76 | 0 | 0.95 | AAGAAAAAGAAAAAGAAAAT |
| 8792~8855 | 29 | 2.2 | 29 | 76 | 13 | 42 | 4 | 1.54 | TAGATATTCTATATTAATATGATTAATAT |
| 15594~15618 | 12 | 2.1 | 12 | 100 | 0 | 48 | 0 | 1.46 | TTAATGATAAGA |
| 28266~28325 | 26 | 2.3 | 26 | 79 | 0 | 45 | 5 | 1.68 | TAGATATAATATAAGGAATCTAAGTA |
| 29086~29123 | 19 | 2.1 | 19 | 90 | 9 | 13 | 10 | 1.19 | TTCTATTTGTATATTTTCT |
| 29538~29582 | 16 | 2.8 | 17 | 86 | 6 | 35 | 8 | 1.31 | TATCTATATATTATATA |
| 29680~29706 | 12 | 2.2 | 12 | 100 | 0 | 25 | 40 | 1.82 | ATCCCCGTACTA |
| 32205~32247 | 22 | 1.9 | 23 | 90 | 4 | 46 | 6 | 1.51 | TATATCACTATATATAATATAAG |
| 32366~32405 | 16 | 2.5 | 15 | 84 | 11 | 65 | 2 | 1.35 | TAAATAAAAGTAAAG |
| 32614~32673 | 32 | 1.9 | 32 | 90 | 10 | 55 | 3 | 1.54 | AATTATATAGAAATATCTAAAGAGTAAAGAGA |
| 60864~60890 | 13 | 2.1 | 13 | 100 | 0 | 44 | 0 | 0.99 | TTTATAATTATAA |
| 67021~67053 | 16 | 2.1 | 16 | 94 | 0 | 45 | 6 | 1.61 | TAAATAGAAATTCTTA |
| 68765~68812 | 15 | 3.1 | 15 | 82 | 17 | 75 | 0 | 1.06 | AAATAAGAATAAAAG |
| 69904~69933 | 15 | 2.0 | 15 | 93 | 0 | 63 | 13 | 1.50 | AAGGAATAAACAAAC |
| 69940~69983 | 21 | 2.1 | 21 | 82 | 0 | 34 | 22 | 1.86 | TAAATCCAAACAACCTTTTCG |
| 73727~73767 | 14 | 3.2 | 13 | 78 | 21 | 21 | 7 | 1.11 | TTTCTTATTTATA |
| 74208~74252 | 13 | 3.5 | 13 | 87 | 6 | 26 | 8 | 1.23 | TCTTATTTTTATA |
| 85176~85230 | 29 | 1.9 | 29 | 82 | 10 | 18 | 10 | 1.15 | TTATTCTAATTTCATTTCTATTTTTATAT |
| 102724~102778 | 14 | 4.2 | 13 | 84 | 11 | 18 | 12 | 1.32 | TTCTTTTCTATTA |
| 102740~102784 | 14 | 3.1 | 14 | 84 | 12 | 24 | 8 | 1.34 | TTCTATTATTATTA |
| 102757~102802 | 17 | 2.6 | 17 | 86 | 3 | 32 | 4 | 1.28 | TATTATTATTATTATAT |
| 116140~116210 | 23 | 2.8 | 24 | 81 | 14 | 33 | 8 | 1.38 | TTATTATATATATATCTTATTCTA |
| 116166~116223 | 15 | 4.1 | 15 | 73 | 20 | 34 | 8 | 1.41 | TTATATACTATATAC |
| 116212~116253 | 21 | 2.1 | 20 | 82 | 8 | 26 | 11 | 1.30 | TACTATTTACTTTACTACTT |
| 116357~116388 | 16 | 2.1 | 15 | 94 | 5 | 56 | 0 | 0.99 | AAATATATTAATATT |
| 130373~130409 | 18 | 2.1 | 18 | 100 | 0 | 16 | 21 | 1.59 | TCTCCAGTTTTTTCATTA |
| 143005~143048 | 17 | 2.5 | 17 | 85 | 3 | 61 | 2 | 1.28 | TAATAATAAGAATAGAA |
| 143029~143083 | 14 | 4.2 | 13 | 82 | 13 | 67 | 1 | 1.32 | AATAATAGAAAAG |
表4 细叶石仙桃叶绿体基因组中串联重复序列
Table 4 Tandem repeat sequences of chloroplast genome in P. cantonensis
位置 Position (bp) | 片段 Fragment (bp) | 重复Copy | 一致 大小 Consensus size (bp) | 匹配比率 Percent matches (%) | 插入删除比率Percent indels (%) | 比例 Percent (%) | 熵 Entropy (0~2) | 一致性序列 Consistent sequence | |
|---|---|---|---|---|---|---|---|---|---|
| A | C | ||||||||
| 3690~3724 | 17 | 2.1 | 17 | 88 | 0 | 8 | 11 | 1.31 | ATTTTCTTTTCCTTGTT |
| 4759~4802 | 18 | 2.4 | 18 | 85 | 7 | 75 | 2 | 1.03 | AAGAAGAAGAAGAAAAAA |
| 4766~4808 | 20 | 2.2 | 20 | 87 | 8 | 76 | 0 | 0.95 | AAGAAAAAGAAAAAGAAAAT |
| 8792~8855 | 29 | 2.2 | 29 | 76 | 13 | 42 | 4 | 1.54 | TAGATATTCTATATTAATATGATTAATAT |
| 15594~15618 | 12 | 2.1 | 12 | 100 | 0 | 48 | 0 | 1.46 | TTAATGATAAGA |
| 28266~28325 | 26 | 2.3 | 26 | 79 | 0 | 45 | 5 | 1.68 | TAGATATAATATAAGGAATCTAAGTA |
| 29086~29123 | 19 | 2.1 | 19 | 90 | 9 | 13 | 10 | 1.19 | TTCTATTTGTATATTTTCT |
| 29538~29582 | 16 | 2.8 | 17 | 86 | 6 | 35 | 8 | 1.31 | TATCTATATATTATATA |
| 29680~29706 | 12 | 2.2 | 12 | 100 | 0 | 25 | 40 | 1.82 | ATCCCCGTACTA |
| 32205~32247 | 22 | 1.9 | 23 | 90 | 4 | 46 | 6 | 1.51 | TATATCACTATATATAATATAAG |
| 32366~32405 | 16 | 2.5 | 15 | 84 | 11 | 65 | 2 | 1.35 | TAAATAAAAGTAAAG |
| 32614~32673 | 32 | 1.9 | 32 | 90 | 10 | 55 | 3 | 1.54 | AATTATATAGAAATATCTAAAGAGTAAAGAGA |
| 60864~60890 | 13 | 2.1 | 13 | 100 | 0 | 44 | 0 | 0.99 | TTTATAATTATAA |
| 67021~67053 | 16 | 2.1 | 16 | 94 | 0 | 45 | 6 | 1.61 | TAAATAGAAATTCTTA |
| 68765~68812 | 15 | 3.1 | 15 | 82 | 17 | 75 | 0 | 1.06 | AAATAAGAATAAAAG |
| 69904~69933 | 15 | 2.0 | 15 | 93 | 0 | 63 | 13 | 1.50 | AAGGAATAAACAAAC |
| 69940~69983 | 21 | 2.1 | 21 | 82 | 0 | 34 | 22 | 1.86 | TAAATCCAAACAACCTTTTCG |
| 73727~73767 | 14 | 3.2 | 13 | 78 | 21 | 21 | 7 | 1.11 | TTTCTTATTTATA |
| 74208~74252 | 13 | 3.5 | 13 | 87 | 6 | 26 | 8 | 1.23 | TCTTATTTTTATA |
| 85176~85230 | 29 | 1.9 | 29 | 82 | 10 | 18 | 10 | 1.15 | TTATTCTAATTTCATTTCTATTTTTATAT |
| 102724~102778 | 14 | 4.2 | 13 | 84 | 11 | 18 | 12 | 1.32 | TTCTTTTCTATTA |
| 102740~102784 | 14 | 3.1 | 14 | 84 | 12 | 24 | 8 | 1.34 | TTCTATTATTATTA |
| 102757~102802 | 17 | 2.6 | 17 | 86 | 3 | 32 | 4 | 1.28 | TATTATTATTATTATAT |
| 116140~116210 | 23 | 2.8 | 24 | 81 | 14 | 33 | 8 | 1.38 | TTATTATATATATATCTTATTCTA |
| 116166~116223 | 15 | 4.1 | 15 | 73 | 20 | 34 | 8 | 1.41 | TTATATACTATATAC |
| 116212~116253 | 21 | 2.1 | 20 | 82 | 8 | 26 | 11 | 1.30 | TACTATTTACTTTACTACTT |
| 116357~116388 | 16 | 2.1 | 15 | 94 | 5 | 56 | 0 | 0.99 | AAATATATTAATATT |
| 130373~130409 | 18 | 2.1 | 18 | 100 | 0 | 16 | 21 | 1.59 | TCTCCAGTTTTTTCATTA |
| 143005~143048 | 17 | 2.5 | 17 | 85 | 3 | 61 | 2 | 1.28 | TAATAATAAGAATAGAA |
| 143029~143083 | 14 | 4.2 | 13 | 82 | 13 | 67 | 1 | 1.32 | AATAATAGAAAAG |
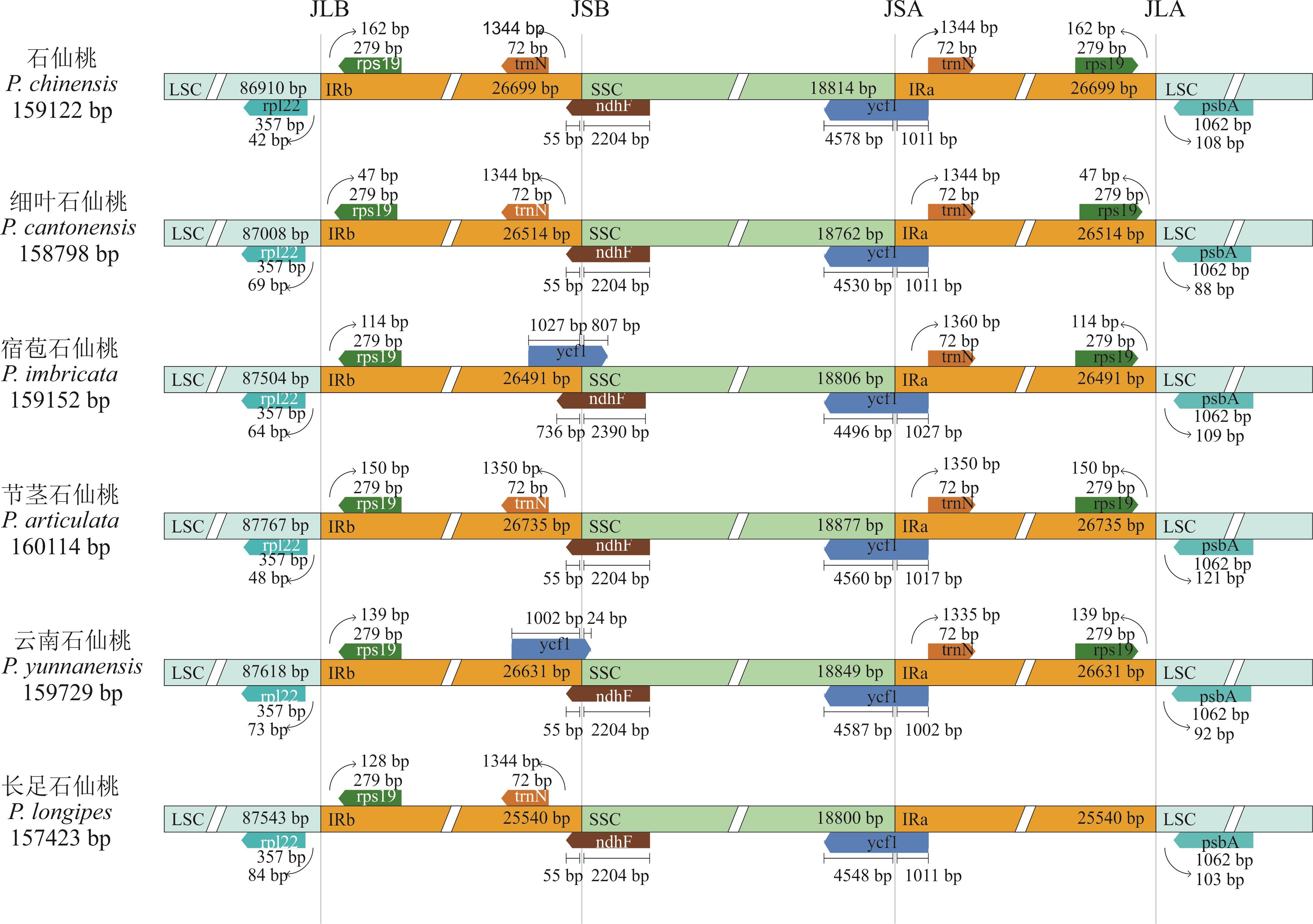
图4 石仙桃属6个物种叶绿体基因组四分体边界LSC: 大单拷贝区Large single-copy; SSC: 小单拷贝区Small single-copy; IRa/b: 2个反向重复区Inverted repeat a/b; JLB(IRb/LSC)、JSB(IRb/SSC)、JSA(SSC/IRa)、 JLA(IRa/LSC): 基因组中每个相应区域之间的连接点Denote the junction sites between each corresponding region in the genome.
Fig.4 The chloroplast genome boundary analysis of six species from Pholidota
| 1 | Chen X, Liu Z, Zhu G, et al. Flora of China. Beijing: Sciences Press, 2009: 335-339. |
| 2 | Liu B, Chen J, Zhang W, et al. The gastrodin biosynthetic pathway in Pholidota chinensis Lindl. revealed by transcriptome and metabolome profiling. Frontiers in Plant Science, 2022,13(11): 1-16. |
| 3 | Wang Y, Dong X J, Zhang M, et al. Advances in the study of the chemincal composition and pharmacological effects of plants of Pholidota chinensis. Research of Zhuang and Yao Ethnic Medicine, 2021(1): 46-51, 182. |
| 王颖, 董雪静, 张淼, 等. 壮药石仙桃的化学成分及药理作用的研究进展. 壮瑶药研究, 2021(1): 46-51, 182. | |
| 4 | Li D L, Zheng X L, Duan L, et al. Ethnobotanical survey of herbal tea plants from the traditional markets in Chaoshan, China. Journal of Ethnopharmacology, 2017,205(9): 195-206. |
| 5 | Wu X C, Liang S, Li Z Y. Advances in chemical constituents and pharmacological activity of Pholidota chinensis Lindl. Chinese Journal of Ethnomedicine and Ethnopharmacy, 2020, 29(4): 60-62. |
| 吴秀彩, 梁爽, 粟正英. 石仙桃属植物化学成分和药理活性的研究概况. 中国民族民间医药, 2020,29(4): 60-62. | |
| 6 | Luo D, Wang Z, Li Z, et al. Structure of an entangled heteropolysaccharide from Pholidota chinensis Lindl and its antioxidant and anti-cancer properties. International Journal of Biological Macromolecules, 2018,112(6): 921-928. |
| 7 | Liu L, Zou M, Zeng K, et al. Chemical constituents and their antioxidant, anti-inflammatory and anti-acetylcholinesterase activities from Pholidota cantonensis. Plant Foods for Human Nutrition, 2021,76(1): 105-110. |
| 8 | LeThi Y, Muniba K, Joonho P. Comparative analysis of chloroplast genome of Desmodium stryacifolium with closely related legume genome from the phaseoloid clade. International Journal of Molecular Sciences, 2023,24(7): 1-13. |
| 9 | Niu Y, Su T, Wu C, et al. Complete chloroplast genome sequences of the medicinal plant Aconitum transsectum (Ranunculaceae): Comparative analysis and phylogenetic relationships. BMC Genomics, 2023,24(1): 1-15. |
| 10 | Zhang G J, Hu Y, Huang W C, et al. Comprehensive phylogenetic analyses of Orchidaceae using nuclear genes and evolutionary insights into epiphytism. Journal of Integrative Plant Biology, 2023,65(5): 1204-1225. |
| 11 | Carla A R, Toscano B A L, Mauad A V S, et al. Phylogenetic position of Centroglossa and Dunstervillea (Ornithocephalus clade: Oncidiinae: Orchidaceae) based on molecular and morphological data. Systematic Botany, 2022,47(4): 927-937. |
| 12 | Liu L N, Zhou G Y, Shen A R, et al. Mycena subpiligera sp. nov., a symbiotic species from China associated with the seed germination of Gastrodia elata. Mycobiology, 2022,50(5): 294-301. |
| 13 | Wu H, Ma P, Li H, et al. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae). Plant Diversity, 2021, 43(1): 15-26. |
| 14 | Bolger A M, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 2014,30(15): 2114-2120. |
| 15 | Jin J J, Yu W B, Yang J B, et al. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology, 2020,21(1): 1-31. |
| 16 | Shi L C, Chen H M, Jiang M, et al. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Research, 2019,47(W1): 65-73. |
| 17 | Kurtz S, Choudhuri J V, Ohlebusch E, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research, 2001,29(22): 4633-4642. |
| 18 | Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Research, 1999,27(2): 573-580. |
| 19 | Beier S, Thiel T, Munch T, et al. MISA-web: A web server for microsatellite prediction. Bioinformatics, 2017,33(16): 2583-2585. |
| 20 | Li H, Guo Q, Xu L, et al. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. Peer Journal Life and Environment, 2023, 11(5): 1-11. |
| 21 | Frazer K A, Pachter L, Poliakov A, et al. VISTA: Computational tools for comparative genomics. Nucleic Acids Research, 2004,32(4): 273-279. |
| 22 | Darling A C, Mau B, Blattner F R, et al. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Research, 2004,14(7): 1394-1403. |
| 23 | Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 2016,33(7): 1870-1874. |
| 24 | Peruzzi L. Advances in plant taxonomy and systematics. Biology, 2023,12(4): 570-573. |
| 25 | Allen J F, Paula W B, Puthiyaveetil S, et al. A structural phylogenetic map for chloroplast photosynthesis. Trends in Plant Science, 2011,16(12): 645-655. |
| 26 | Rao W H, Huang J, Li L Q, et al. The complete chloroplast genome of Pholidota imbricata (Orchidaceae). Mitochondrial DNA Part B, 2019,4(2): 3523-3524. |
| 27 | Li C R, Luo Y, Zhao P, et al. The complete chloroplast genome sequence of Pholidota articulata (Orchidaceae), a rarely medicinal orchid. Mitochondrial DNA Part B, 2020,5(4): 3801-3802. |
| 28 | Xie Z N, Lao J, Liu H, et al. Characterization of the chloroplast genome of medicinal herb Polygonatum cyrtonema and identification of molecular markers by comparative analysis. Genome, 2023,66(4): 80-90. |
| 29 | Zhang X, Zhang G, Jiang Y, et al. Complete chloroplast genome sequence of endangered species in the genus opisthopappus C. Shih: Characterization, species identification, and phylogenetic relationships. Genes, 2022,13(12): 2410-2452. |
| 30 | Park J, Suh Y, Kim S. A complete chloroplast genome sequence of Gastrodia elata (Orchidaceae) represents high sequence variation in the species. Mitochondrial DNA Part B, 2020,5(1): 517-519. |
| 31 | Cai Z, Wang H, Wang G. Complete chloroplast genome sequence of Bletilla striata (Thunb.) Reichb. f., a Chinese folk medicinal plant. Mitochondrial DNA Part B, 2020,5(3): 2239-2240. |
| 32 | Xue Q Q, Yang J P, Yu W H, et al. The climate changes promoted the chloroplast genomic evolution of Dendrobium orchids among multiple photosynthetic pathways. BMC Plant Biology, 2023,23(1): 189-202. |
| 33 | Deng H, Zhang L S, Zhang G Q, et al. Evolutionary history of PEPC genes in green plants: Implications for the evolution of CAM in orchids. Molecular Phylogenetics and Evolution, 2016,94(Part B): 559-564. |
| 34 | Yoko Y, Masamitsu O, Yuto H, et al. Photosynthesis and leaf structure of F1 hybrids between Cymbidium ensifolium (C3) and C. bicolor subsp. pubescens (CAM). Annals of Botany, 2022, 12: 157. |
| [1] | 刘昊, 李显炀, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 紫花苜蓿SAUR基因家族的鉴定及其在非生物胁迫中的表达模式研究[J]. 草业学报, 2024, 33(4): 135-153. |
| [2] | 黄祥, 何梦瑶, 王子煊, 楚光明, 江萍. 紫叶风箱果叶绿体基因组特征及绣线菊亚科系统发育分析[J]. 草业学报, 2024, 33(3): 161-173. |
| [3] | 黄丽娟, 孙镕基, 高文婧, 张志飞, 陈桂华. 全株水稻表面优势乳酸菌的筛选与鉴定[J]. 草业学报, 2024, 33(1): 117-125. |
| [4] | 吕自立, 刘彬, 常凤, 马紫荆, 曹秋梅. 巴音布鲁克高寒草甸物种多样性与系统发育多样性沿海拔梯度分布格局及驱动因子[J]. 草业学报, 2023, 32(7): 12-22. |
| [5] | 于晓东, 余浩洋, 杨旭, 赵东旭, 张林刚. 内蒙古两种生态型羊草叶绿体基因组序列差异分析[J]. 草业学报, 2023, 32(7): 72-84. |
| [6] | 高守舆, 李钰莹, 杨志青, 董宽虎, 夏方山. 白羊草叶绿体基因组密码子使用偏好性分析[J]. 草业学报, 2023, 32(7): 85-95. |
| [7] | 李艳鹏, 魏娜, 翟庆妍, 李杭, 张吉宇, 刘文献. 全基因组水平白花草木樨TCP基因家族的鉴定及在干旱胁迫下表达模式分析[J]. 草业学报, 2023, 32(4): 101-111. |
| [8] | 张国香, 郭卫冷, 毕铭钰, 张力爽, 王丹, 郭长虹. 紫花苜蓿CAX基因家族鉴定及其对非生物胁迫的响应分析[J]. 草业学报, 2022, 31(12): 106-117. |
| [9] | 刘涛, 刘玉萍, 富贵, 吕婷, 刘峰, 张雨, 苏丹丹, 王亚男, 郑长远, 苏旭. 基于流式细胞法和K-mer分析法检测沙鞭基因组大小[J]. 草业学报, 2022, 31(12): 133-145. |
| [10] | 魏娜, 李艳鹏, 马艺桐, 刘文献. 全基因组水平紫花苜蓿TCP基因家族的鉴定及其在干旱胁迫下表达模式分析[J]. 草业学报, 2022, 31(1): 118-130. |
| [11] | 王艺蒙, 马甜甜, 欧阳子凤, 张吉宇. 无芒隐子草全基因组水平全长LTR反转录转座子鉴定及其中断基因分析[J]. 草业学报, 2021, 30(5): 121-133. |
| [12] | 纪会, 官久强, 王会, 周建旭, 阿农呷, 何宗伟, 樊珍详, 邱龙康, 曹诗晓, 安添午, 柏琴, 钟金城, 罗晓林. 亚丁牦牛和拉日马牦牛遗传多样性及遗传结构分析[J]. 草业学报, 2021, 30(5): 134-145. |
| [13] | 刘文文, 崔会婷, 尉春雪, 龙瑞才, 康俊梅, 杨青川, 王珍. 蒺藜苜蓿叶绿素酸酯a加氧酶(MtCAO)基因的克隆与功能分析[J]. 草业学报, 2020, 29(5): 171-181. |
| [14] | 李淑洁, 张金文, 裴怀弟, 王红梅, 罗俊杰, 林玉红. 兰州百合同源四倍体诱导及其基因组大小估算[J]. 草业学报, 2020, 29(3): 190-196. |
| [15] | 刘淑娟, 张晓军, 李欣, 刘成, 白建荣, 任永康, 郑军, 李世姣, 郭慧娟, 梅超, 张树伟, 畅志坚, 乔麟轶. 中间偃麦草第6同源群特异STS标记开发[J]. 草业学报, 2019, 28(4): 139-145. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||