

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2026, Vol. 35 ›› Issue (2): 195-207.DOI: 10.11686/cyxb2025190
王召明1( ), 郑丽娜1, 张跃华1, 赵韦1, 陈翔1,2, 贾振宇1(
), 郑丽娜1, 张跃华1, 赵韦1, 陈翔1,2, 贾振宇1( )
)
收稿日期:2025-05-09
修回日期:2025-06-11
出版日期:2026-02-20
发布日期:2025-12-24
通讯作者:
贾振宇
作者简介:Corresponding author. E-mail: Jzhenyu0612@163.com基金资助:
Zhao-ming WANG1( ), Li-na ZHENG1, Yue-hua ZHANG1, Wei ZHAO1, Xiang CHEN1,2, Zhen-yu JIA1(
), Li-na ZHENG1, Yue-hua ZHANG1, Wei ZHAO1, Xiang CHEN1,2, Zhen-yu JIA1( )
)
Received:2025-05-09
Revised:2025-06-11
Online:2026-02-20
Published:2025-12-24
Contact:
Zhen-yu JIA
摘要:
植物生长与抗逆表现依赖于植物根与共生微生物之间的有益互作。低秋眠级苜蓿在短日照和低温条件下适应性强,但其根系相关微生物组装特征及其品种间差异尚不明确。本研究依托甘肃甘南野外试验站,种植4种低秋眠级苜蓿品种:兰苜1号、公农1号、金皇后(前3种均为紫花苜蓿)和甘农1号(杂花苜蓿)。利用16S rRNA基因扩增子测序,系统分析4个苜蓿品种根际与根内细菌群落特征及组装机制。结果表明,苜蓿根际细菌群落物种多样性显著高于根内,不同品种之间根系细菌群落结构同样差异显著。与其他品种相比,新型耐寒品系兰苜1号在根际特异性富集了链嗜酸菌属,并在一定程度上提高了根系富集的细菌群落物种多样性,增强了微生物共现网络的复杂性和连通性。群落构建过程分析表明,所有品种的根际和根内细菌群落组装主要受到异质性选择(42%~55%)和匀质性选择(56%~83%)过程影响,其中兰苜1号、公农1号和甘农1号品种根系细菌群落相比于金皇后受到更大程度的确定性选择过程影响。本研究揭示了低秋眠级苜蓿根际与根内微生物群落的组装模式及品种间差异,为调控植物微生物组以提升寒区牧草适应性提供理论依据。
王召明, 郑丽娜, 张跃华, 赵韦, 陈翔, 贾振宇. 低秋眠耐寒苜蓿根系微生物组装特征及其品种间差异[J]. 草业学报, 2026, 35(2): 195-207.
Zhao-ming WANG, Li-na ZHENG, Yue-hua ZHANG, Wei ZHAO, Xiang CHEN, Zhen-yu JIA. Bacterial community assembly in the rhizosphere and endosphere of different perennial alfalfa varieties with low fall dormancy rates[J]. Acta Prataculturae Sinica, 2026, 35(2): 195-207.

图2 不同苜蓿品种的根际与根内微生物多样性差异(a、b、c)及门水平物种组成(d)不同苜蓿品种包括:甘农1号(Gan1)、公农1号(Gong1)、金皇后(GE)与兰苜1号(Lan1)。下同。Different alfalfa varieties include: Gannong No.1 (Gan1), Gongnong No.1 (Gong1), Golden empress (GE) and Lanmu No.1 (Lan1). The same below.
Fig.2 Differences in rhizosphere and endosphere microbial richness (a, b, c) and phylum level species composition (d) among different alfalfa varieties
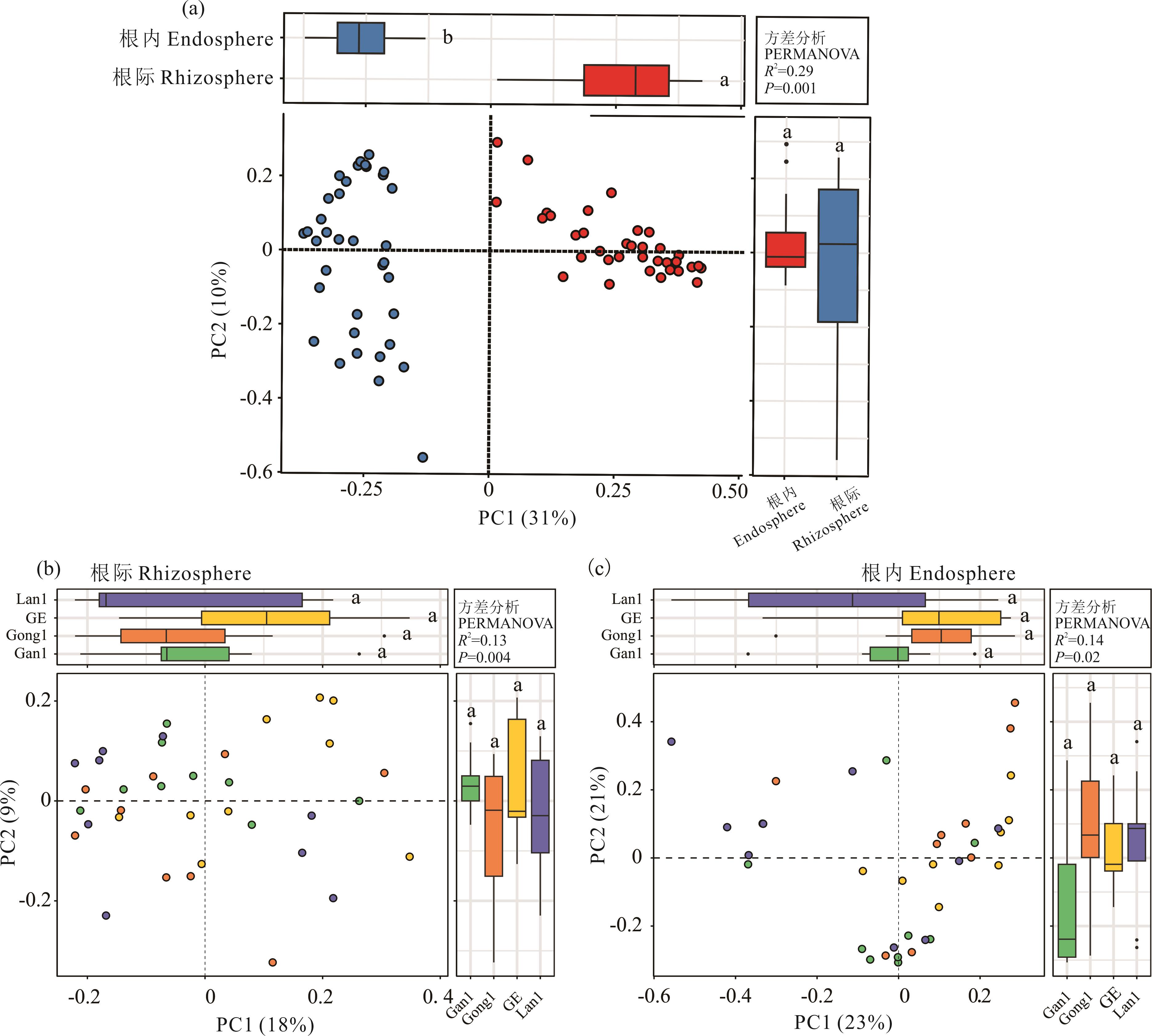
图3 不同苜蓿品种的根际与根内微生物beta多样性分析(a): 根际与根内细菌群落的整体差异 Overall differences in rhizosphere vs endosphere bacterial communities; (b): 品种间根际细菌群落的差异 Differences in rhizosphere bacterial communities among varieties; (c): 品种间根内细菌群落的差异 Differences in endophytic bacterial communities among varieties.
Fig. 3 Beta diversity of rhizosphere and endophytic microorganisms in different alfalfa varieties

图4 不同苜蓿品种的根际与根内丰度存在显著差异的微生物数量及根内富集的微生物种类(a)、(b)、(c)、(d): 不同苜蓿品种根际-根内细菌物种富集分析Enrichment analysis of rhizosphere and endosphere bacterial species in different varieties; (e): 根内富集的细菌物种的系统发育关系及其在不同苜蓿品种中的分布模式Phylogenetic relationships of bacterial species enriched in endosphere and their distribution in different varieties.
Fig.4 Number of microorganisms with significant differences in rhizosphere and endosphere abundance among alfalfa varieties and the taxonomy of microorganisms enriched in endosphere
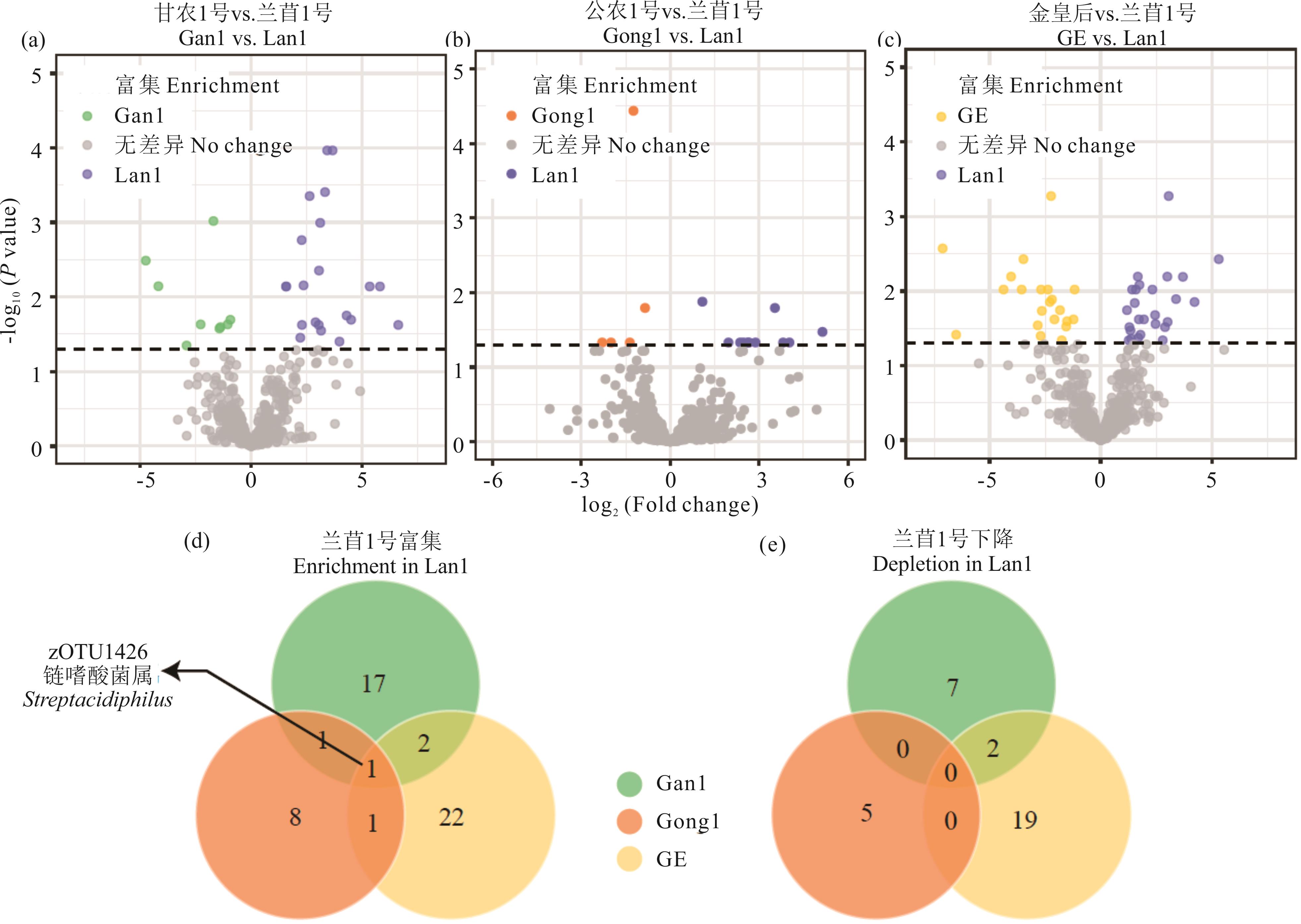
图5 兰苜1号(Lan1)与其他品种根际微生物差异分析(a)、(b)、(c)分别是甘农1号、公农1号、金皇后与兰苜1号相比的根际细菌丰度差异分析The differences in rhizosphere bacterial abundance between Gan1 and Lan1, between Gong1 and Lan1, and between GE and Lan1, respectively; (d): 兰苜1号根际相比于其他品种富集的细菌种类数The number of bacterial species enriched in the rhizosphere of Lan1 compared to other varieties; (e): 兰苜1号根际相比于其他品种消减的细菌种类数The number of bacterial species reduced in the rhizosphere of Lan1 compared to other varieties.
Fig.5 Differential analysis of rhizosphere microorganisms between Lanmu No.1 (Lan1) and other varieties
| [1] | Shi S, Nuccio E E, Shi Z J, et al. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecology Letters, 2016, 19(8): 926-936. |
| [2] | Ruan Y, Wang D S, Guo S W, et al. Fingerprints of carbon metabolisms of gourd as rootstock and watermelon as scion in rhizosphere of different types of soils-characteristics and differences. Acta Pedologica Sinica, 2018, 55(4): 967-976. |
| 阮杨, 王东升, 郭世伟, 等. 葫芦与西瓜砧穗在不同土壤上根际碳代谢指纹特征与差异. 土壤学报, 2018, 55(4): 967-976. | |
| [3] | Kuzyakov Y, Razavi B S. Rhizosphere size and shape: Temporal dynamics and spatial stationarity. Soil Biology and Biochemistry, 2019, 135(9): 343-360. |
| [4] | Nuccio E E, Starr E, Karaoz U, et al. Niche differentiation is spatially and temporally regulated in the rhizosphere. The ISME Journal, 2020, 14(4): 999-1014. |
| [5] | Ravanbakhsh M, Kowalchuk G A, Jousset A. Root-associated microorganisms reprogram plant life history along the growth–stress resistance tradeoff. The ISME Journal, 2019, 13(12): 3093-3101. |
| [6] | Edwards J, Johnson C, Santos-Medellín C, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proceedings of the National Academy of Sciences, 2015, 112(8): 911-920. |
| [7] | Ruan Y, Wang T T, Guo S W, et al. Plant grafting shapes complexity and co-occurrence of rhizobacterial assemblages. Microbial Ecology, 2020, 80(8): 643-655. |
| [8] | Ruan Y, Wang T T, Guo S W, et al. Chimeric plants favor asynchrony of conditionally rare bacterial species facilitating functional complementarity in rhizosphere. Biology and Fertility of Soils, 2022, 58(4): 459-470. |
| [9] | Li B, Li Y Y, Wu H M, et al. Root exudates drive interspecific facilitation by enhancing nodulation and n2 fixation. Proceedings of the National Academy of Sciences, 2016, 113(23): 6496-6501. |
| [10] | Liu Q, Dai H C, Cheng H, et al. Rhizosphere-associated bacterial and fungal communities of two maize hybrids under increased nitrogen fertilization. Frontiers in Plant Science, 2025, 16: 1549995. |
| [11] | Hou M, Leng C, Zhu J, et al. Alpine and subalpine plant microbiome mediated plants adapt to the cold environment: A systematic review. Environmental Microbiome, 2024, 19(1): 82. |
| [12] | Lan H, Gorfer M, Otgonsuren B, et al. Growth characteristics and freezing tolerance of ectomycorrhizal and saprotrophic fungi: responses to normal and freezing temperatures. Forests, 2025, 16(2): 191. |
| [13] | Zhou J Z, Ning D L. Stochastic community assembly: Does it matter in microbial ecology? Microbiology and Molecular Biology Reviews, 2017, 81(4): 1-32. |
| [14] | Sasse J, Martinoia E, Northen T. Feed your friends: Do plant exudates shape the root microbiome? Trends in Plant Science, 2018, 23(1): 25-41. |
| [15] | Wang T T, Ruan Y, Xu Q C, et al. Effect of plant-derived microbial soil legacy in a grafting system—a turn for the better. Microbiome, 2024, 12(1): 234. |
| [16] | Li H, Wang J, Liu Q, et al. Effects of consecutive monoculture of sweet potato on soil bacterial community as determined by pyrosequencing. Journal of Basic Microbiology, 2019, 59(2): 181-191. |
| [17] | Edgar R C. Search and clustering orders of magnitude faster than blast. Bioinformatics, 2010, 26(19): 2460-2461. |
| [18] | Ruan Y, Ling N, Jiang S J, et al. Warming and altered precipitation independently and interactively suppress alpine soil microbial growth in a decadal-long experiment. eLife, 2024, 12: 1-17. |
| [19] | Kong Y L, Ling N, Xue C, et al. Long-term fertilization regimes change soil nitrification potential by impacting active autotrophic ammonia oxidizers and nitrite oxidizers as assessed by DNA stable isotope probing. Environmental Microbiology, 2019, 21(4): 1224-1240. |
| [20] | Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 2008, 9(1): 559. |
| [21] | Yang X, Li X F, Xie G Q, et al. Experimental construction plan for network data mining based on igraph. Computer Knowledge and Technology, 2019, 15(21): 29-31. |
| 杨雄, 李晓芳, 谢光前, 等. 基于igraph的网络数据挖掘实验建设方案. 电脑知识与技术, 2019, 15(21): 29-31. | |
| [22] | Feng K, Zhang Z J, Cai W W, et al. Biodiversity and species competition regulate the resilience of microbial biofilm community. Molecular Ecology, 2017, 26(21): 6170-6182. |
| [23] | Feng Y, Chen R, Stegen J C, et al. Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Molecular Ecology, 2018, 27(24): 5238-5251. |
| [24] | Ling N, Song Y, Raza W, et al. The response of root-associated bacterial community to the grafting of watermelon. Plant and Soil, 2015, 391(1): 253-264. |
| [25] | Wang J, Jiang L B, Wu R L. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytologist, 2017, 214(1): 56-65. |
| [26] | Gaion L A, Trevisan B L, Carvalho R F. Grafting in vegetable crops: A great technique for agriculture. International Journal of Vegetable Science, 2018, 24(1): 85-102. |
| [27] | Maynard D S, Crowther T W, Bradford M A. Competitive network determines the direction of the diversity-function relationship. Proceedings of the National Academy of Sciences, 2017, 114(43): 11464-11469. |
| [28] | Konopka A, Lindemann S, Fredrickson J. Dynamics in microbial communities: Unraveling mechanisms to identify principles. The ISME Journal, 2015, 9(7): 1488-1495. |
| [29] | Wei Z, Yang T, Friman V P, et al. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nature Communications, 2015, 6(1): 8413. |
| [30] | van Elsas J D, Chiurazzi M, Mallon C A, et al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proceedings of the National Academy of Sciences, 2012, 109(4): 1159-1164. |
| [31] | Song Y, Zhu C, Raza W, et al. Coupling of the chemical niche and microbiome in the rhizosphere: Implications from watermelon grafting. Frontiers of Agricultural Science and Engineering, 2016, 3(3): 249-262. |
| [32] | Chen X, Wu J H, Yang F J, et al. New insight into the mechanism by which antifreeze peptides regulate the physiological function of Streptococcus thermophilus subjected to freezing stress. Journal of Advanced Research, 2023, 45(5): 127-140. |
| [33] | Berendsen R L, van Verk M C, Stringlis I A, et al. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS 374 and WCS 417. BMC Genomics, 2015, 16(539): 1-23. |
| [34] | Chatterjee P, Samaddar S, Anandham R, et al. Beneficial soil bacterium Pseudomonas frederiksbergensis os261 augments salt tolerance and promotes red pepper plant growth. Frontiers in Plant Science, 2017, 8: 705. |
| [35] | Subramanian P, Kim K, Krishnamoorthy R, et al. Cold stress tolerance in psychrotolerant soil bacteria and their conferred chilling resistance in tomato (Solanum lycopersicum Mill.) under low temperatures. PLoS One, 2016, 11(8): e0161592. |
| [36] | Matteo C, Alessandra S D F, Stefania D, et al. Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytologist, 2018, 220(4): 1296-1308. |
| [37] | Layeghifard M, Hwang D M, Guttman D S. Disentangling interactions in the microbiome: A network perspective. Trends in Microbiology, 2017, 25(3): 217-228. |
| [38] | Toju H, Okayasu K, Notaguchi M. Leaf-associated microbiomes of grafted tomato plants. Scientific Reports, 2019, 9(1): 1787. |
| [39] | Song Y, Ling N, Ma J H, et al. Grafting resulted in a distinct proteomic profile of watermelon root exudates relative to the un-grafted watermelon and the rootstock plant. Journal of Plant Growth Regulation, 2016, 35(3): 778-791. |
| [40] | Wagg C, Schlaeppi K, Banerjee S, et al. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nature Communications, 2019, 10(1): 4841. |
| [41] | Peng G S, Wu J. Optimal network topology for structural robustness based on natural connectivity. Physica A: Statistical Mechanics and Its Applications, 2016, 443: 212-220. |
| [42] | Hassan S, Mathesius U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. Journal of Experimental Botany, 2012, 63(9): 3429-3444. |
| [43] | He D X, Singh S K, Peng L, et al. Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. The ISME Journal, 2022, 16(11): 2622-2632. |
| [44] | Attia S, Russel J, Mortensen M S, et al. Unexpected diversity among small-scale sample replicates of defined plant root compartments. The ISME Journal, 2022, 16(4): 997-1003. |
| [45] | Fukami T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annual Review of Ecology Evolution and Systematics, 2015, 46(1): 1-23. |
| [46] | Chase J M. Community assembly: When should history matter? Oecologia, 2003, 136(4): 489-498. |
| [47] | Doornbos R F, Loon L C V, Bakker P A H M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agronomy for Sustainable Development, 2012, 32(1): 227-243. |
| [1] | 宋一欣, 李明源, 麦日艳古·亚生, 王继莲. 新疆高寒草地3种植物根际土壤真菌群落结构及功能多样性[J]. 草业学报, 2026, 35(2): 167-178. |
| [2] | 马红钰, 周小国, 王宝, 宋渝川, 艾克热木·阿不拉提江null, 蒋邵丽, 闵九洲, 赵红梅, 程军回. 准噶尔荒漠梭梭和柽柳根际土壤微生物功能基因丰度变化特征[J]. 草业学报, 2025, 34(8): 109-122. |
| [3] | 汤珊珊, 胡敏. 禾本科植物根际土壤酶活性和细菌群落结构差异[J]. 草业学报, 2025, 34(8): 99-108. |
| [4] | 王颖, 李明源, 麦日艳古·亚生null, 王继莲. 新疆托木尔峰不同植物根际土壤真菌群落结构比较研究[J]. 草业学报, 2025, 34(7): 83-94. |
| [5] | 邓文辉, 宋珂辰, 张浩, 管思雨, 雍嘉仪, 胡海英. 降水变化条件下荒漠草原优势植物根际微生物群落结构和多样性特征研究[J]. 草业学报, 2025, 34(5): 12-26. |
| [6] | 陶惠赟, 杨闰艳, 李延灿, 刘亚鹏, 祁鹤兴. 黄河源区矮嵩草根际土壤微生物多样性及对环境因子的响应[J]. 草业学报, 2025, 34(12): 16-32. |
| [7] | 李春艳, 王钱进, 周芯合, 曹文静, 赵梦丽, 虞方伯. Burkholderia sp. SX9菌剂对白三叶生长和改良土壤的影响[J]. 草业学报, 2025, 34(11): 53-65. |
| [8] | 伍国强, 于祖隆, 魏明. PGPR调控植物响应逆境胁迫的作用机制[J]. 草业学报, 2024, 33(6): 203-218. |
| [9] | 何升然, 刘晓静, 赵雅姣, 汪雪, 王静. 紫花苜蓿/甜高粱间作对根际土壤特性及微生物群落特征的影响[J]. 草业学报, 2024, 33(5): 92-105. |
| [10] | 程鑫宇, 王继莲, 麦日艳古·亚生null, 李明源. 盐爪爪根际土壤产IAA菌株分离及促生特性分析[J]. 草业学报, 2024, 33(4): 110-121. |
| [11] | 马源, 王晓丽, 马玉寿, 张德罡. 高寒草甸退化程度对优势物种根际土壤真菌群落和生态网络的影响[J]. 草业学报, 2024, 33(2): 125-137. |
| [12] | 邢静, 范文强, 王佳妮, 石凤翎. 干旱胁迫下 2个扁蓿豆品种根际细菌多样性及土壤灭菌对其生长的影响[J]. 草业学报, 2024, 33(12): 147-159. |
| [13] | 许代香, 杨建峰, 苏杭, 翟建荣, 綦才, 赵龙刚, 郭彦军. 间作模式下作物根际土壤代谢物对微生物群落的影响[J]. 草业学报, 2023, 32(11): 65-80. |
| [14] | 苗阳阳, 张艳蕊, 宋标, 刘旭桐, 张安琪, 吕金泽, 张浩, 张小华, 欧阳佳慧, 李旺, 曲善民. 碱蓬根际和内生细菌菌株对盐碱胁迫下苜蓿生长的影响[J]. 草业学报, 2022, 31(9): 107-117. |
| [15] | 刘晓婷, 姚拓. 高寒草地耐低温植物根际促生菌的筛选鉴定及特性研究[J]. 草业学报, 2022, 31(8): 178-187. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||