

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (7): 53-67.DOI: 10.11686/cyxb2023334
吴毅1( ), 冯雅岚2, 王添宁1, 琚吉浩1, 肖慧淑1, 马超1, 张均1(
), 冯雅岚2, 王添宁1, 琚吉浩1, 肖慧淑1, 马超1, 张均1( )
)
收稿日期:2023-09-14
修回日期:2023-11-15
出版日期:2024-07-20
发布日期:2024-04-08
通讯作者:
张均
作者简介:E-mail: zhangjun0105@126.com基金资助:
Yi WU1( ), Ya-lan FENG2, Tian-ning WANG1, Ji-hao JU1, Hui-shu XIAO1, Chao MA1, Jun ZHANG1(
), Ya-lan FENG2, Tian-ning WANG1, Ji-hao JU1, Hui-shu XIAO1, Chao MA1, Jun ZHANG1( )
)
Received:2023-09-14
Revised:2023-11-15
Online:2024-07-20
Published:2024-04-08
Contact:
Jun ZHANG
摘要:
热激蛋白70(heat shock protein 70, Hsp70)在植物发育过程以及响应生物和非生物胁迫中起着重要作用。为探究小麦Hsp70基因家族进化关系、功能以及表达模式,本研究对乌拉尔图小麦、拟斯卑尔脱山羊草、二粒小麦、粗山羊草以及普通小麦的Hsp70基因进行全面的生物信息学分析,并通过RT-qPCR方法分析其部分Hsp70基因在不同外源激素和环境胁迫条件下的表达模式。结果表明,从乌拉尔图小麦、拟斯卑尔脱山羊草、二粒小麦、粗山羊草和普通小麦5个物种中,分别鉴定出30、41、60、28和94个Hsp70基因;系统发育分析表明5个物种Hsp70家族成员分为5个亚家族组,每组成员数量不相等,其中大部分成员分布在第Ⅰ组,且同一亚家族中大多数的Hsp70成员具有相似的基因结构和保守基序;进一步综合分析5个物种Hsp70基因的染色体定位和重复事件,发现Hsp70基因在5个物种的各染色体上分布不均匀,此外从5个物种中共发现12个串联重复事件和110个片段复制事件,表明片段复制事件促进了小麦Hsp70基因家族的扩张;顺式作用元件分析表明,5个物种Hsp70基因的启动子区域存在多种光响应元件、逆境响应元件、激素响应元件以及生长发育调节元件;此外RT-qPCR结果表明,5个物种部分Hsp70基因在不同激素处理和逆境胁迫下具有不同程度的响应,在高温和干旱胁迫下,所选8个Hsp70基因均上调表达。小麦及其祖先物种Hsp70基因的鉴定及其进化过程为进一步研究Hsp70基因在小麦生长发育过程中的功能以及在逆境胁迫下的响应机制提供理论基础。
吴毅, 冯雅岚, 王添宁, 琚吉浩, 肖慧淑, 马超, 张均. 小麦及其祖先物种Hsp70基因家族鉴定与表达分析[J]. 草业学报, 2024, 33(7): 53-67.
Yi WU, Ya-lan FENG, Tian-ning WANG, Ji-hao JU, Hui-shu XIAO, Chao MA, Jun ZHANG. Genome-wide identification and expression analysis of the Hsp70 gene family in wheat and its ancestral species[J]. Acta Prataculturae Sinica, 2024, 33(7): 53-67.
| 基因号Gene ID | 基因名Gene name | 正向引物Forward primer (5’-3’) | 反向引物Reverse primer (5’-3’) |
|---|---|---|---|
| TraesCS5A02G479300.1 | TaHsp70-54 | GCCACAGCTGGTGACACTCAC | TGGAATAGAAATCAACTCCCTCG |
| TraesCS5B02G087700.1 | TaHsp70-57 | CATCAGTGGCAACCCGAGAG | GAAGTCAATGCCCTCAAACAGC |
| TraesCS5D02G492900.1 | TaHsp70-68 | GGTGGCACTTTTGATGTCTCC | CCAAGATGAGTGTCACCAGCC |
| TRITD5Av1G040700.2 | TtHsp70-36 | GGACTCTCTCCTCCACTGCG | TGGACAGCAGCCCCGTAG |
| TRITD5Bv1G039950.1 | TtHsp70-44 | GCAAAGATGGACAAGAGCACC | TGGACAGCAGCCCCGTAC |
| XP_048532408.1 | TuHsp70-18 | GCCACAGCTGGTGACACTCAC | TGGAATAGAAATCAACTCCCTCG |
| AespeY2032CH5S01G103600.1 | AesHsp70-25 | CATCAGTGGCAACCCGAGAG | GAAGTCAATGCCCTCAAACAGC |
| XP_020200619.1 | AetHsp70-20 | GGTGGCACTTTTGATGTCTCC | CCAAGATGAGTGTCACCAGCC |
| TraesCS1B02G283900.1 | Actin | GTTCCAATCTATGAGGGATACACGC | GAACCTCCACTGAGAACAACATTACC |
表1 部分Hsp70基因荧光定量所用的引物
Table 1 Some primers for RT-qPCR of Hsp70 genes
| 基因号Gene ID | 基因名Gene name | 正向引物Forward primer (5’-3’) | 反向引物Reverse primer (5’-3’) |
|---|---|---|---|
| TraesCS5A02G479300.1 | TaHsp70-54 | GCCACAGCTGGTGACACTCAC | TGGAATAGAAATCAACTCCCTCG |
| TraesCS5B02G087700.1 | TaHsp70-57 | CATCAGTGGCAACCCGAGAG | GAAGTCAATGCCCTCAAACAGC |
| TraesCS5D02G492900.1 | TaHsp70-68 | GGTGGCACTTTTGATGTCTCC | CCAAGATGAGTGTCACCAGCC |
| TRITD5Av1G040700.2 | TtHsp70-36 | GGACTCTCTCCTCCACTGCG | TGGACAGCAGCCCCGTAG |
| TRITD5Bv1G039950.1 | TtHsp70-44 | GCAAAGATGGACAAGAGCACC | TGGACAGCAGCCCCGTAC |
| XP_048532408.1 | TuHsp70-18 | GCCACAGCTGGTGACACTCAC | TGGAATAGAAATCAACTCCCTCG |
| AespeY2032CH5S01G103600.1 | AesHsp70-25 | CATCAGTGGCAACCCGAGAG | GAAGTCAATGCCCTCAAACAGC |
| XP_020200619.1 | AetHsp70-20 | GGTGGCACTTTTGATGTCTCC | CCAAGATGAGTGTCACCAGCC |
| TraesCS1B02G283900.1 | Actin | GTTCCAATCTATGAGGGATACACGC | GAACCTCCACTGAGAACAACATTACC |

图3 小麦及其祖先物种Hsp70基因的保守基序及基因结构分析A: Hsp70基因家族的系统进化树 Phylogenetic tree of the Hsp70 family; B: Hsp70基因家族的保守基序 Conserved motifs of the Hsp70 family; C: Hsp70基因家族的基因结构 Gene structure of the Hsp70 family.CDS: 编码区Coding sequences; UTR: 非翻译区Untranslated regions.
Fig.3 Analysis of the phylogenetic relationship and gene structure of Hsp70 gene in wheat and its ancestral species

图4 小麦及其祖先物种Hsp70基因的染色体分布染色体中条带颜色的深浅表示基因密度的大小。The depth of band color in chromosomes indicates the size of gene density.
Fig.4 Chromosomal distribution of Hsp70 gene in wheat and its ancestral species

图5 小麦及其祖先物种Hsp70基因的染色体位置及共线性关系所有共线性区域和基因由灰线连接,红线表示Hsp70基因的片段重复基因对。All syntenic blocks and genes are linked by the grey lines, and red lines represent the fragment-repeat gene pair of the Hsp70 genes.
Fig.5 Chromosomal positions and synteny relationships of Hsp70 genes in wheat and its ancestral species
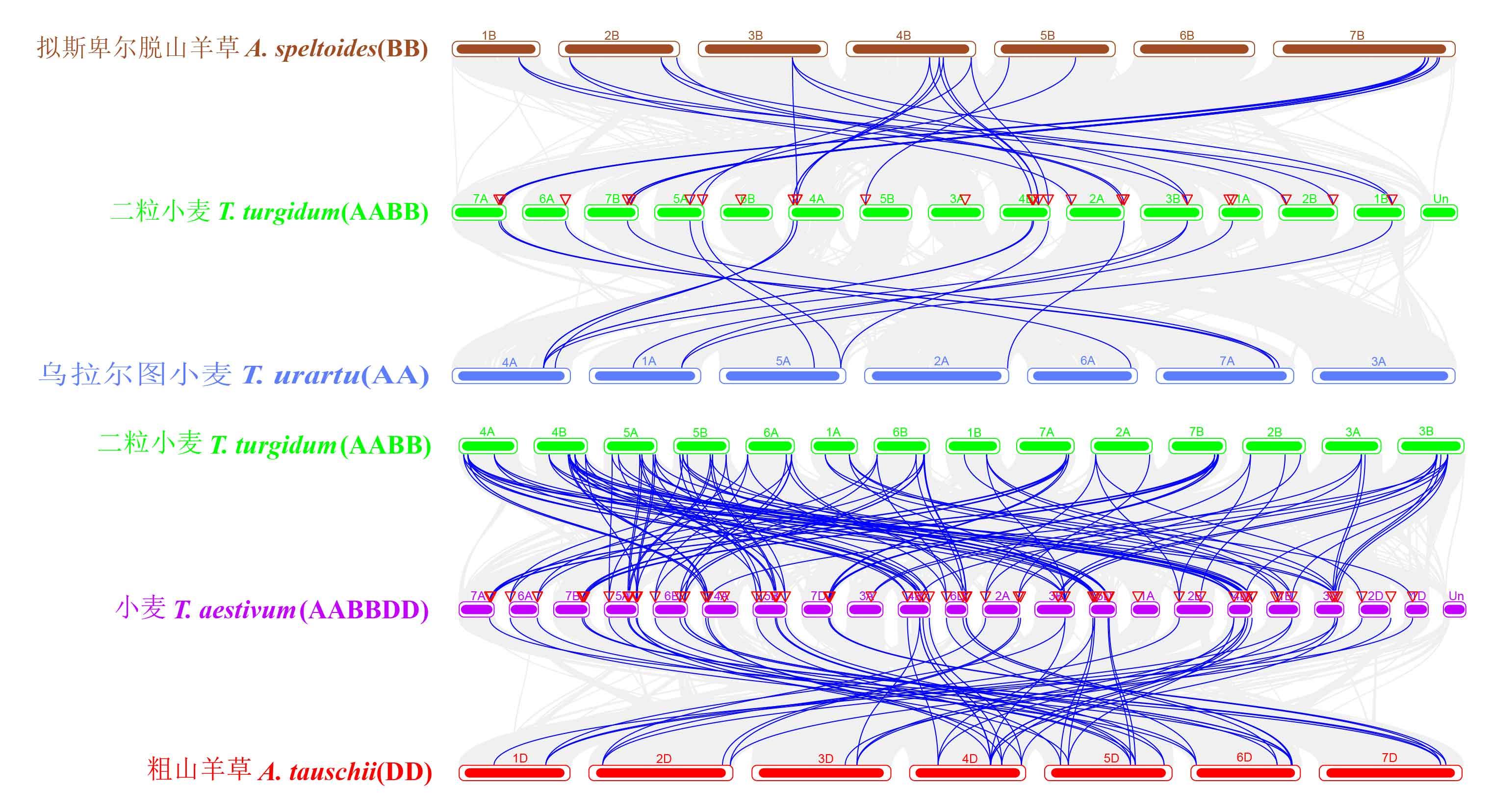
图6 小麦及其祖先物种Hsp70基因的同源性分析背景中的灰色线表示两个基因组内的共线块,蓝色线突出显示共线的Hsp70基因对。Gray lines in the background indicate the collinear blocks within two genomes, while blue lines highlight the syntenic Hsp70 gene pairs.
Fig.6 Homologous analysis of the Hsp70 genes in wheat and its ancestral species

图7 小麦及其祖先物种Hsp70基因启动子顺式作用元件不同的启动子由不同颜色的色块表示。Different promoters are represented by blocks of different colors.
Fig.7 Promoter cis-regulatory element of the Hsp70 genes in wheat and its ancestral species
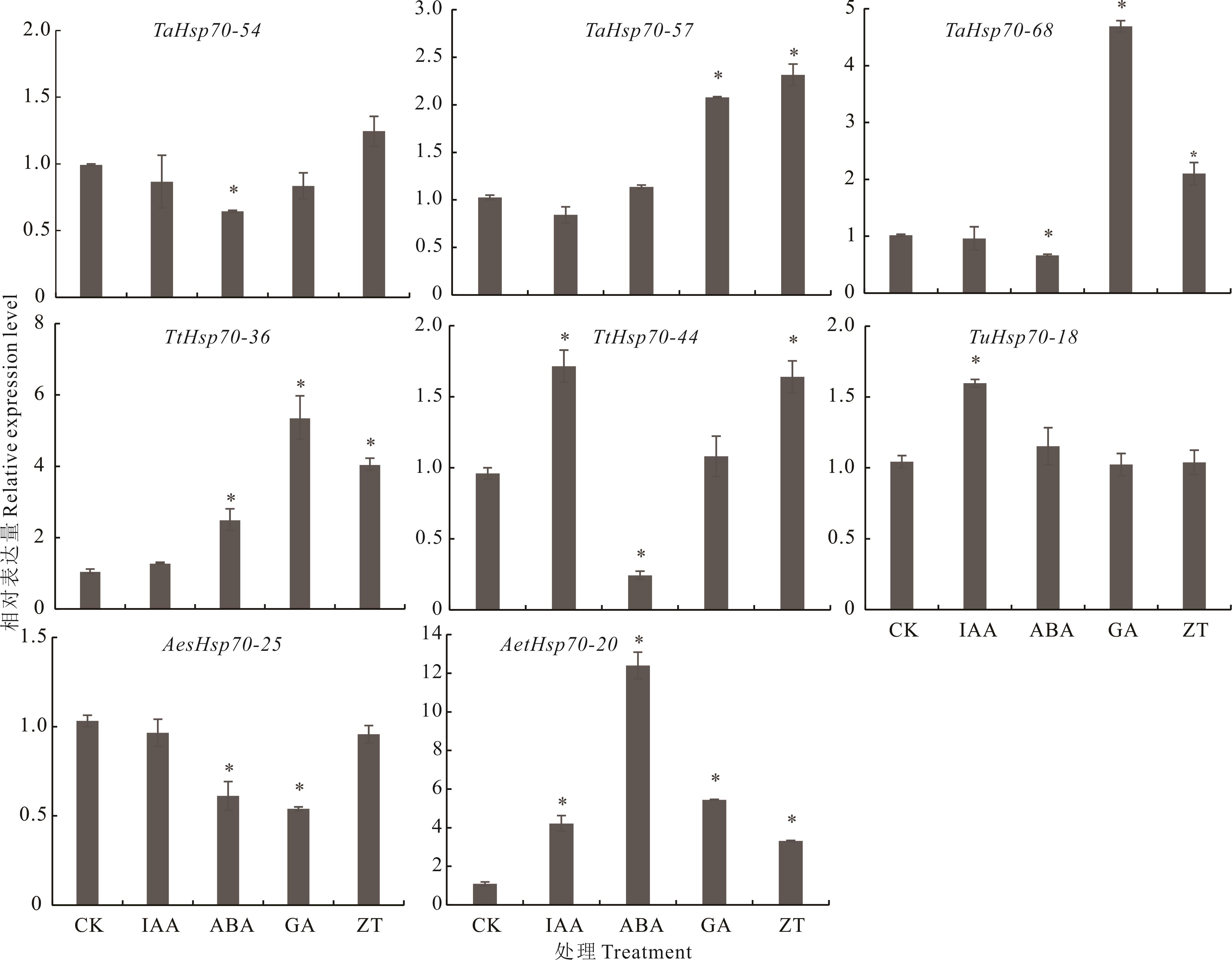
图8 Hsp70基因家族成员在4种激素处理下的表达模式分析CK: 对照Control; IAA: 生长素Auxin; ABA: 脱落酸Abscisic acid; GA: 赤霉素Gibberellin; ZT: 玉米素Zeatin; *: P<0.05. 下同The same below.
Fig.8 Expression pattern analysis of Hsp70 gene family members under four hormone treatments

图9 Hsp70基因家族成员在4种逆境胁迫下的表达模式分析CK: 对照Control; S: 盐Salting; LT: 低温Low temperature; HT:高温High temperature; D:干旱Drought.
Fig.9 Expression patterns analysis of Hsp70 gene family members under four kinds of stress
| 1 | Park C J, Seo Y S. Heat shock proteins: A review of the molecular chaperones for plant immunity. The Plant Pathology Journal, 2015, 31(4): 323-333. |
| 2 | Haq U S, Khan A, Ali M, et al. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences, 2019, 20(21): 5321. |
| 3 | Boston R S, Viitanen P V, Vierling E. Molecular chaperones and protein folding in plants. Plant Molecular Biology, 1996, 32(1/2): 191-222. |
| 4 | Wang W X, Vinocur B, Shoseyov O, et al. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science, 2004, 9(5): 244-252. |
| 5 | Kiang J G, Tsokos G C. Heat shock protein 70 kDa: Molecular biology, biochemistry, and physiology. Pharmacology and Therapeutics, 1998, 80(2): 183-201. |
| 6 | Dragovic Z, Broadley S A, Shomura Y, et al. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. The EMBO Journal, 2006, 25(11): 2519-2528. |
| 7 | Hartl F U, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature, 2011, 475(7356): 324-332. |
| 8 | Lin B L, Wang J S, Liu H C, et al. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress and Chaperones, 2001, 6(3): 201-208. |
| 9 | Sarkar N K, Sharma P, Grover A. Functional analysis of HSP70 superfamily proteins of rice (Oryza sativa). Cell Stress and Chaperones, 2013, 18(4): 427-437. |
| 10 | Du Q L, Jiang J M, Chen M Q, et al. Cloning, expression analysis and prokaryotic expression of heat shock protein HSP70 gene in rice. Journal of Plant Protection, 2021, 48(3): 620-629. |
| 杜巧丽, 蒋君梅, 陈美晴, 等. 水稻热休克蛋白HSP70基因克隆、表达分析及原核表达. 植物保护学报, 2021, 48(3): 620-629. | |
| 11 | Zhang L, Zhao H K, Dong Q L, et al. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Frontiers in Plant Science, 2015, 6: 773. |
| 12 | Rehman A, Atif R M, Qayyum A, et al. Genome-wide identification and characterization of HSP70 gene family in four species of cotton. Genomics, 2020, 112(6): 4442-4453. |
| 13 | Song J H, Ma H L, Weng Q Y, et al. Genome-wide identification and analysis of HSP70 gene family in maize. Journal of Nuclear Agricultural Sciences, 2017, 31(7): 1245-1254. |
| 宋晋辉, 马海莲, 瓮巧云, 等. 玉米HSP70基因家族的全基因组鉴定与分析. 核农学报, 2017, 31(7): 1245-1254. | |
| 14 | El Baidouri M, Murat F, Veyssiere M, et al. Reconciling the evolutionary origin of bread wheat (Triticum aestivum). The New Phytologist, 2017, 213(3): 1477-1486. |
| 15 | Maccaferri M, Harris N S, Twardziok S O, et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nature Genetics, 2019, 51(5): 885-895. |
| 16 | Cheng H, Liu J, Wen J, et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biology, 2019, 20(1): 136. |
| 17 | Gardiner L J, Wingen L U, Bailey P, et al. Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biology, 2019, 20(1): 69. |
| 18 | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| 19 | Bailey T L, Johnson J, Grant C E, et al. The MEME suite. Nucleic Acids Research, 2015, 43(1): 39-49. |
| 20 | Chen C J, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 21 | Wang Y P, Tang H B, Debarry J D, et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 2012, 40(7): e49. |
| 22 | Kang C H, Jung W Y, Kang Y H, et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death and Differentiation, 2006, 13(1): 84-95. |
| 23 | Chen E L, Fan Z Y, Wang S F, et al. Bioinformatics of tobacco (Nicotiana tabacum) Hsp70 gene family and expression analysis of NtHsp70Chl in midrib. Chinese Tobacco Science, 2018, 39(2): 8-16. |
| 陈二龙, 范志勇, 王松峰, 等. 烟草Hsp70基因家族的鉴定及NtHsp70Chl基因的表达分析. 中国烟草科学, 2018, 39(2): 8-16. | |
| 24 | Liu J, Pang X, Cheng Y, et al. The Hsp70 gene family in solanum tuberosum: Genome-wide identification, phylogeny, and expression patterns. Scientific Reports, 2018, 8(1): 16628. |
| 25 | Song G, Fang Z G, Wang Y L, et al. Genome-wide identification and bioinformatics analysis of Hsp70 family genes in switchgrass. Pratacultural Science, 2022, 39(10): 2112-2126. |
| 宋刚, 方志刚, 王玉龙, 等. 柳枝稷Hsp70家族基因鉴定与生物信息学分析. 草业科学, 2022, 39(10): 2112-2126. | |
| 26 | Zhang G W, Liu L L, Wang X R, et al. Genome-wide identification and bioinformatics analysis of HSP70 genes in foxtail millet. Acta Agriculturae Zhejiangensis, 2015, 27(7): 1127-1133. |
| 张古文, 刘莉莉, 王显瑞, 等. 谷子HSP70基因家族的全基因组鉴定及生物信息学分析. 浙江农业学报, 2015, 27(7): 1127-1133. | |
| 27 | Wang X S, Jin Z, Ding Y N, et al. Characterization of HSP70 family in watermelon (Citrullus lanatus): Identification, structure, evolution, and potential function in response to ABA, cold and drought stress. Frontiers in Genetics, 2023, 14: 1201535. |
| 28 | de Souza S J, Long M, Gilbert W. Introns and gene evolution. Genes Cells, 1996, 1(6): 493-505. |
| 29 | Babenko V N, Rogozin I B, Mekhedov S L, et al. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Research, 2004, 32(12): 3724-3733. |
| 30 | Roy S W, Penny D. On the incidence of intron loss and gain in paralogous gene families. Molecular Biology and Evolution, 2007, 24(8): 1579-1581. |
| 31 | Sémon M, Wolfe K H. Consequences of genome duplication. Current Opinion in Genetics and Development, 2007, 17(6): 505-512. |
| 32 | Lawton-Rauh A. Evolutionary dynamics of duplicated genes in plants. Molecular Phylogenetics and Evolution, 2003, 29(3): 396-409. |
| 33 | Magadum S, Banerjee U, Murugan P, et al. Gene duplication as a major force in evolution. Journal of Genetics, 2013, 92(1): 155-161. |
| 34 | Zhang Y, Zheng L J, Yun L, et al. Catalase (CAT) gene family in wheat (Triticum aestivum L.): Evolution, expression pattern and function analysis. International Journal of Molecular Sciences, 2022, 23(1): 542. |
| 35 | Su P H, Li H M. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiology, 2008, 146(3): 1231-1241. |
| 36 | Sable A, Rai K M, Choudhary A, et al. Inhibition of heat shock proteins HSP90 and HSP70 induce oxidative stress, suppressing cotton fiber development. Scientific Reports, 2018, 8(1): 3620. |
| 37 | He Z S, Xie R, Wang Y Z, et al. Cloning and characterization of a heat shock protein 70 gene, MsHSP70-1, in Medicago sativa. Acta Biochimica et Biophysica Sinica, 2008, 40(3): 209-216. |
| 38 | Kurepa J, Wang S H, Li Y, et al. Proteasome regulation, plant growth and stress tolerance. Plant Signaling and Behavior, 2009, 4(10): 924-927. |
| [1] | 徐寿霞. 基于meta分析的丛枝菌根对小麦产量和品质的影响[J]. 草业学报, 2024, 33(7): 192-204. |
| [2] | 张俊豪, 柴雪茹, 马嵩科, 张冬霞, 张静, 乔唱唱, 李爽, 黄明, 王贺正. 秸秆还田配施磷肥对豫西旱地小麦碳同化物积累的影响及其生理机制[J]. 草业学报, 2024, 33(6): 89-104. |
| [3] | 孔海明, 宋家兴, 杨静, 李倩, 杨培志, 曹玉曼. 紫花苜蓿CAMTA基因家族鉴定及其在非生物胁迫下的表达模式分析[J]. 草业学报, 2024, 33(5): 143-154. |
| [4] | 刘昊, 李显炀, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 紫花苜蓿SAUR基因家族的鉴定及其在非生物胁迫中的表达模式研究[J]. 草业学报, 2024, 33(4): 135-153. |
| [5] | 李显炀, 刘昊, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 全基因组水平紫花苜蓿WRKY转录因子家族鉴定与表达模式分析[J]. 草业学报, 2024, 33(4): 154-170. |
| [6] | 韩硕, 韩晓文, 胡义锋, 陈中义, 朱永兴, 尹军良. 空心莲子草SOD基因家族鉴定和表达模式分析[J]. 草业学报, 2024, 33(1): 102-116. |
| [7] | 史先飞, 高宇, 黄旭升, 周雅莉, 蔡桂萍, 李昕儒, 李润植, 薛金爱. 油莎豆CeWRKY转录因子响应非生物胁迫的功能表征[J]. 草业学报, 2023, 32(8): 186-201. |
| [8] | 张振粉, 黄荣, 李向阳, 姚博, 赵桂琴. 基于Illumina MiSeq高通量测序的燕麦种带细菌多样性及功能分析[J]. 草业学报, 2023, 32(7): 96-108. |
| [9] | 马嵩科, 霍克, 张冬霞, 张静, 张俊豪, 柴雪茹, 王贺正. 玉米秸秆还田配施氮肥对豫西旱地小麦土壤酶活性和氮肥利用效率的影响[J]. 草业学报, 2023, 32(6): 120-133. |
| [10] | 柴继宽, 琚泽亮, 赵桂琴. 低温和低pH胁迫下青贮乳酸菌OL77的内参基因筛选及CspP基因的表达模式分析[J]. 草业学报, 2023, 32(5): 147-158. |
| [11] | 李艳鹏, 魏娜, 翟庆妍, 李杭, 张吉宇, 刘文献. 全基因组水平白花草木樨TCP基因家族的鉴定及在干旱胁迫下表达模式分析[J]. 草业学报, 2023, 32(4): 101-111. |
| [12] | 刘亚男, 于人杰, 高燕丽, 康俊梅, 杨青川, 武志海, 王珍. 蒺藜苜蓿膜联蛋白MtANN2基因的表达模式及盐胁迫下的功能分析[J]. 草业学报, 2022, 31(5): 124-134. |
| [13] | 常利芳, 李欣, 郭慧娟, 乔麟轶, 张树伟, 陈芳, 畅志坚, 张晓军. 小偃麦衍生系表型遗传多样性分析及综合评价[J]. 草业学报, 2022, 31(11): 61-74. |
| [14] | 周涛, 牟乐, 苏楷淇, 张筠钰, 杨惠敏. 间作比例和调亏灌溉对春小麦/紫花苜蓿间作中春小麦灌浆期旗叶性状的影响[J]. 草业学报, 2022, 31(10): 145-153. |
| [15] | 张丹丹, 张元庆, 程景, 靳光, 李博, 王栋才, 徐芳, 孙锐锋. 不同粗饲料组合对晋南牛瘤胃体外发酵特性的研究[J]. 草业学报, 2021, 30(7): 93-100. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||