

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (5): 143-154.DOI: 10.11686/cyxb2023240
• 研究论文 • 上一篇
孔海明1( ), 宋家兴1, 杨静1, 李倩2, 杨培志1, 曹玉曼1(
), 宋家兴1, 杨静1, 李倩2, 杨培志1, 曹玉曼1( )
)
收稿日期:2023-07-13
修回日期:2023-09-04
出版日期:2024-05-20
发布日期:2024-02-03
通讯作者:
曹玉曼
作者简介:E-mail: yumancao@nwafu.edu.cn基金资助:
Hai-ming KONG1( ), Jia-xing SONG1, Jing YANG1, Qian LI2, Pei-zhi YANG1, Yu-man CAO1(
), Jia-xing SONG1, Jing YANG1, Qian LI2, Pei-zhi YANG1, Yu-man CAO1( )
)
Received:2023-07-13
Revised:2023-09-04
Online:2024-05-20
Published:2024-02-03
Contact:
Yu-man CAO
摘要:
钙调蛋白结合转录激活因子(CAMTA)是一类重要的钙调素结合蛋白,在激素信号转导、发育调控和环境胁迫耐受中发挥着重要作用。本研究采用生物信息学技术,基于紫花苜蓿“新疆大叶”参考基因组,对紫花苜蓿中CAMTA家族成员进行鉴定,并对这些基因的理化性质、系统发育树、保守结构域、染色体上位置、顺式作用元件、转录表达谱进行分析和验证。结果表明,共鉴定出17个MsCAMTA基因,MsCAMTA家族成员可划分为3个亚家族,亚家族成员在基因结构、保守基序位置上较为相似。染色体定位结果显示,MsCAMTA家族成员不均匀地分布在7条染色体上。启动子区具有大量响应低温、盐胁迫及植物激素信号相关的顺式作用元件。此外,采用RT-qPCR对盐(300 mmol·L-1 NaCl)、模拟干旱(400 mmol·L-1 甘露醇)、低温(10 ℃)和脱落酸(100 μmol·L-1)处理下紫花苜蓿叶片中MsCAMTA1、MsCAMTA3、MsCAMTA11和MsCAMTA12的表达模式进行了初步研究。结果表明,4个MsCAMTA候选基因在各胁迫处理下均有一定程度的响应,且在盐胁迫下的表达量都呈上调趋势,表明MsCAMTA基因可能通过整合多种逆境应激信号,参与紫花苜蓿响应各非生物胁迫的应答过程。本研究结果将为进一步探索MsCAMTA基因在植物应对逆境胁迫中的功能提供参考。
孔海明, 宋家兴, 杨静, 李倩, 杨培志, 曹玉曼. 紫花苜蓿CAMTA基因家族鉴定及其在非生物胁迫下的表达模式分析[J]. 草业学报, 2024, 33(5): 143-154.
Hai-ming KONG, Jia-xing SONG, Jing YANG, Qian LI, Pei-zhi YANG, Yu-man CAO. Identification and transcript profiling of the CAMTA gene family under abiotic stress in alfalfa[J]. Acta Prataculturae Sinica, 2024, 33(5): 143-154.
| 基因Gene | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsCAMTA1 | GAGGAGGAAGCACGCCAG | GCGGCTGCCATTGCTTTT |
| MsCAMTA3 | GCGGACAGTTGGCTTCCT | GCCCTTTGTGCCCATTGC |
| MsCAMTA11 | TCTTCATTGGGCGGCGTT | AGCTAGGTCGGCTGGTGT |
| MsCAMTA12 | TGGTGCATCTGCTGGAGC | CCAGTCCCTTATGCCCGC |
| MsActin | GACAATGGAACTGGAATGG | CAATACCGTGCTCAATGG |
表1 RT-qPCR引物信息
Table 1 Primers used for RT-qPCR
| 基因Gene | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsCAMTA1 | GAGGAGGAAGCACGCCAG | GCGGCTGCCATTGCTTTT |
| MsCAMTA3 | GCGGACAGTTGGCTTCCT | GCCCTTTGTGCCCATTGC |
| MsCAMTA11 | TCTTCATTGGGCGGCGTT | AGCTAGGTCGGCTGGTGT |
| MsCAMTA12 | TGGTGCATCTGCTGGAGC | CCAGTCCCTTATGCCCGC |
| MsActin | GACAATGGAACTGGAATGG | CAATACCGTGCTCAATGG |
基因名称 Gene name | 基因号 Gene ID | 染色体 Chromosome | 不稳定 指数 Instability index | 疏水性指数 Grand average of hydrophobicity (GRAVY) | 脂肪指数 Aliphatic index | 蛋白质长度 Protein length (aa) | 分子量 Molecular weight (kDa) | 等电点 Isoelectric point (pI) | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| MsCAMTA1 | MS.gene00191.t1 | Chr2.1 | 42.00 | -0.418 | 79.07 | 915 | 103.86 | 6.49 | 细胞核Nucleus |
| MsCAMTA2 | MS.gene56018.t1 | Chr2.3 | 42.52 | -0.418 | 78.73 | 914 | 103.81 | 6.48 | 细胞核Nucleus |
| MsCAMTA3 | MS.gene002202.t1 | Chr2.3 | 47.33 | -0.522 | 76.66 | 913 | 101.75 | 5.24 | 细胞核Nucleus |
| MsCAMTA4 | MS.gene09226.t1 | Chr4.1 | 47.88 | -0.596 | 69.50 | 960 | 107.40 | 5.44 | 细胞核Nucleus |
| MsCAMTA5 | MS.gene57296.t1 | Chr4.1 | 43.64 | -0.443 | 79.89 | 1745 | 196.21 | 6.12 | 细胞核Nucleus |
| MsCAMTA6 | MS.gene028443.t1 | Chr4.1 | 45.76 | -0.575 | 70.13 | 1029 | 116.11 | 5.70 | 细胞核Nucleus |
| MsCAMTA7 | MS.gene90181.t1 | Chr4.2 | 45.67 | -0.570 | 70.51 | 1029 | 116.11 | 5.67 | 细胞核Nucleus |
| MsCAMTA8 | MS.gene030815.t1 | Chr4.2 | 45.61 | -0.619 | 74.79 | 1086 | 123.35 | 5.77 | 细胞核Nucleus |
| MsCAMTA9 | MS.gene70367.t1 | Chr4.3 | 45.59 | -0.567 | 70.41 | 1029 | 116.08 | 5.73 | 细胞核Nucleus |
| MsCAMTA10 | MS.gene023441.t1 | Chr4.3 | 47.56 | -0.603 | 69.40 | 960 | 107.42 | 5.44 | 细胞核Nucleus |
| MsCAMTA11 | MS.gene061784.t1 | Chr4.3 | 45.61 | -0.619 | 74.79 | 1086 | 123.35 | 5.77 | 细胞核Nucleus |
| MsCAMTA12 | MS.gene09019.t1 | Chr4.4 | 47.56 | -0.603 | 69.40 | 960 | 107.42 | 5.44 | 细胞核Nucleus |
| MsCAMTA13 | MS.gene28071.t1 | Chr4.4 | 44.14 | -0.420 | 81.90 | 1437 | 161.54 | 6.30 | 叶绿体Chloroplast |
| MsCAMTA14 | MS.gene35018.t1 | Chr4.4 | 46.06 | -0.572 | 70.13 | 1029 | 116.16 | 5.70 | 细胞核Nucleus |
| MsCAMTA15 | MS.gene61520.t1 | Chr8.3 | 39.09 | -0.467 | 80.03 | 864 | 98.08 | 7.14 | 细胞核Nucleus |
| MsCAMTA16 | MS.gene61521.t1 | Chr8.3 | 40.02 | -0.474 | 80.42 | 924 | 105.32 | 7.37 | 细胞核Nucleus |
| MsCAMTA17 | MS.gene042294.t1 | Chr8.3 | 39.70 | -0.470 | 80.51 | 923 | 105.16 | 7.15 | 细胞核Nucleus |
表2 紫花苜蓿CAMTA家族成员基本信息
Table 2 Basic information of CAMTA family members in M. sativa
基因名称 Gene name | 基因号 Gene ID | 染色体 Chromosome | 不稳定 指数 Instability index | 疏水性指数 Grand average of hydrophobicity (GRAVY) | 脂肪指数 Aliphatic index | 蛋白质长度 Protein length (aa) | 分子量 Molecular weight (kDa) | 等电点 Isoelectric point (pI) | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| MsCAMTA1 | MS.gene00191.t1 | Chr2.1 | 42.00 | -0.418 | 79.07 | 915 | 103.86 | 6.49 | 细胞核Nucleus |
| MsCAMTA2 | MS.gene56018.t1 | Chr2.3 | 42.52 | -0.418 | 78.73 | 914 | 103.81 | 6.48 | 细胞核Nucleus |
| MsCAMTA3 | MS.gene002202.t1 | Chr2.3 | 47.33 | -0.522 | 76.66 | 913 | 101.75 | 5.24 | 细胞核Nucleus |
| MsCAMTA4 | MS.gene09226.t1 | Chr4.1 | 47.88 | -0.596 | 69.50 | 960 | 107.40 | 5.44 | 细胞核Nucleus |
| MsCAMTA5 | MS.gene57296.t1 | Chr4.1 | 43.64 | -0.443 | 79.89 | 1745 | 196.21 | 6.12 | 细胞核Nucleus |
| MsCAMTA6 | MS.gene028443.t1 | Chr4.1 | 45.76 | -0.575 | 70.13 | 1029 | 116.11 | 5.70 | 细胞核Nucleus |
| MsCAMTA7 | MS.gene90181.t1 | Chr4.2 | 45.67 | -0.570 | 70.51 | 1029 | 116.11 | 5.67 | 细胞核Nucleus |
| MsCAMTA8 | MS.gene030815.t1 | Chr4.2 | 45.61 | -0.619 | 74.79 | 1086 | 123.35 | 5.77 | 细胞核Nucleus |
| MsCAMTA9 | MS.gene70367.t1 | Chr4.3 | 45.59 | -0.567 | 70.41 | 1029 | 116.08 | 5.73 | 细胞核Nucleus |
| MsCAMTA10 | MS.gene023441.t1 | Chr4.3 | 47.56 | -0.603 | 69.40 | 960 | 107.42 | 5.44 | 细胞核Nucleus |
| MsCAMTA11 | MS.gene061784.t1 | Chr4.3 | 45.61 | -0.619 | 74.79 | 1086 | 123.35 | 5.77 | 细胞核Nucleus |
| MsCAMTA12 | MS.gene09019.t1 | Chr4.4 | 47.56 | -0.603 | 69.40 | 960 | 107.42 | 5.44 | 细胞核Nucleus |
| MsCAMTA13 | MS.gene28071.t1 | Chr4.4 | 44.14 | -0.420 | 81.90 | 1437 | 161.54 | 6.30 | 叶绿体Chloroplast |
| MsCAMTA14 | MS.gene35018.t1 | Chr4.4 | 46.06 | -0.572 | 70.13 | 1029 | 116.16 | 5.70 | 细胞核Nucleus |
| MsCAMTA15 | MS.gene61520.t1 | Chr8.3 | 39.09 | -0.467 | 80.03 | 864 | 98.08 | 7.14 | 细胞核Nucleus |
| MsCAMTA16 | MS.gene61521.t1 | Chr8.3 | 40.02 | -0.474 | 80.42 | 924 | 105.32 | 7.37 | 细胞核Nucleus |
| MsCAMTA17 | MS.gene042294.t1 | Chr8.3 | 39.70 | -0.470 | 80.51 | 923 | 105.16 | 7.15 | 细胞核Nucleus |
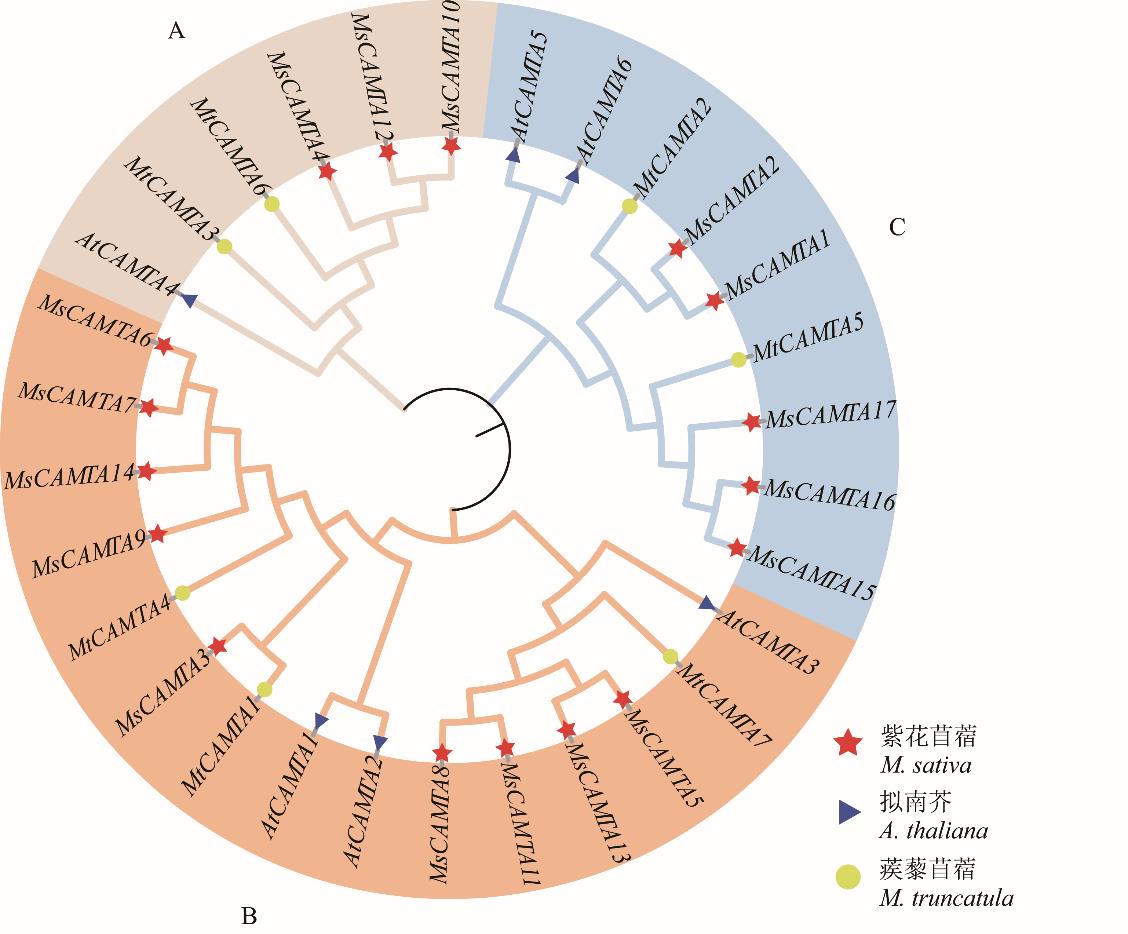
图1 基于紫花苜蓿、拟南芥和蒺藜苜蓿CAMTA氨基酸序列构建系统进化树
Fig.1 Construction of phylogenetic tree based on CAMTA amino acid sequences of M. sativa, A.thaliana and M. truncatula

图2 MsCAMTA的进化树和保守motif分布(A)、基因结构(B)和motif序列(C)字母越大,表示该位置越保守。The larger the letter, the more conservative the position is. 1~10: 基序1~10 Motif 1-10.
Fig.2 The phylogenetic tree, distribution and characteristics of conserved motifs (A), gene structure (B) and motif sequences of MsCAMTA
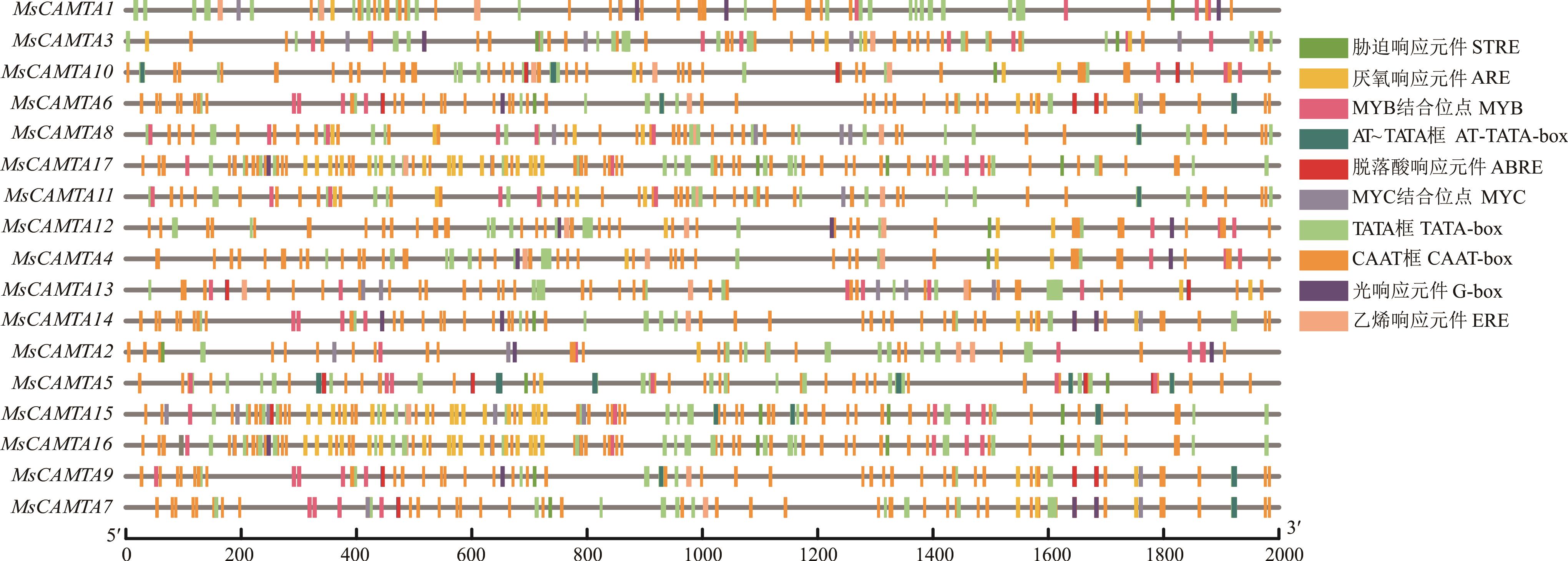
图4 紫花苜蓿CAMTA家族成员启动子区域顺式作用元件STRE: 胁迫响应元件Stress responsive element; ARE: 厌氧响应元件Anaerobic induction responsive element; MYB: MYB结合位点MYB binding site; ABRE: 脱落酸响应元件Abscisic acid responsive element; MYC: MYC结合位点MYC binding site; ERE: 乙烯响应元件Ethylene responsive element.
Fig.4 Cis-regulatory element in promoter region of CAMTA family genes in alfalfa
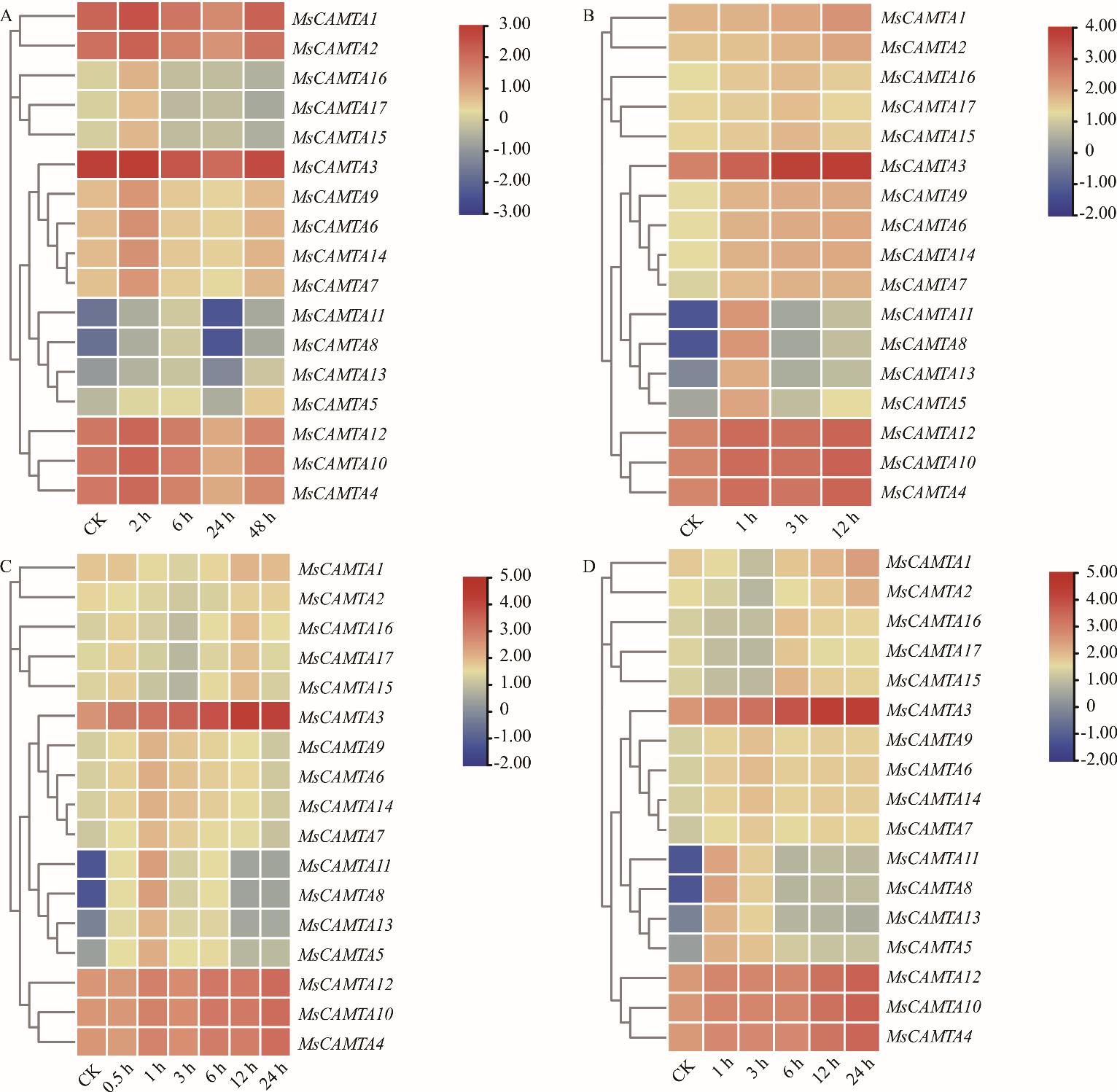
图5 MsCAMTA基因的表达模式A: 冷处理Cold treatment; B: 10 mol·L-1 ABA处理10 mol·L-1 ABA treatment; C: 250 mmol·L-1盐处理250 mmol·L-1 NaCl treatment; D: 400 mmol·L-1甘露醇处理400 mmol·L-1 mannitol treament. 表达谱中用蓝、黄、红3种颜色表示基因表达强度,红色越深说明信号越强,蓝色越深说明信号越弱,黄色代表中等水平。Three colors (blue, yellow, red) are used in the expression spectrum to indicate gene expression intensity. The darker the red color, the stronger the signal. The darker the blue color, the weaker the signal. Yellow represents a moderate level.
Fig.5 Expression patterns of MsCAMTA genes
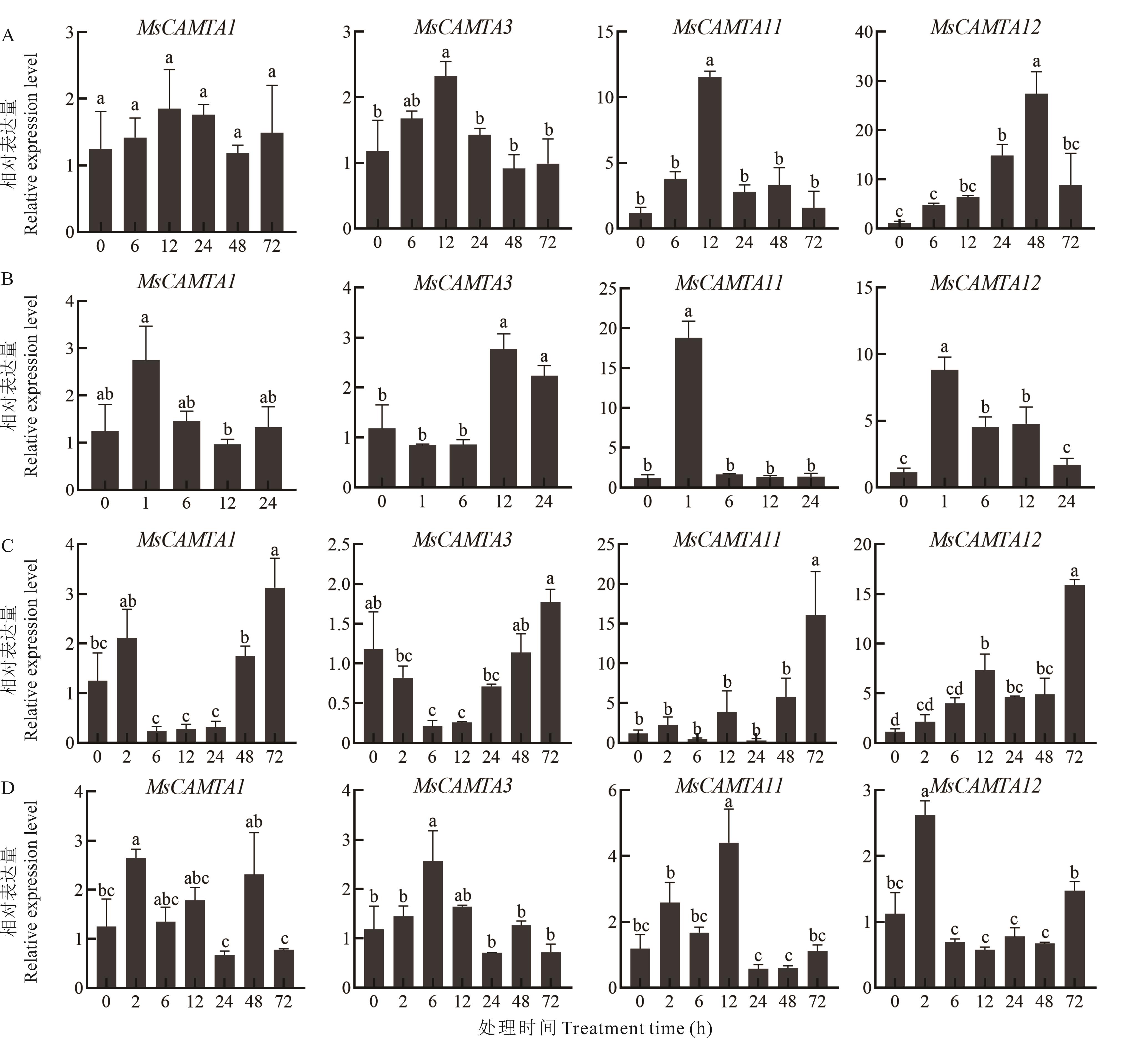
图6 紫花苜蓿CAMTA基因家族在胁迫下的RT-qPCR表达分析A: 冷处理Cold treatment; B: 100 μmol·L-1 ABA处理100 μmol·L-1 ABA treatment; C: 300 mmol·L-1盐处理300 mmol·L-1 NaCl treatment; D: 400 mmol·L-1甘露醇处理400 mmol·L-1 mannitol treament. 数据是3次生物学重复的平均值,不同小写字母表示在0.05水平存在显著性差异。The data is the average of three biological repetitions, the different lowercase letters indicate significant differences at the 0.05 level.
Fig.6 RT-qPCR expression analysis of CAMTA gene family under abiotic stress in alfalfa
| 1 | Galon Y, Snir O, Fromm H. How calmodulin binding transcription activators (CAMTAs) mediate auxin responses. Plant Signaling Behavior, 2010, 5(10): 1311-1314. |
| 2 | Reddy A S N, Ali G S, Celesnik H, et al. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. The Plant Cell, 2011, 23(6): 2010-2032. |
| 3 | Xiao P X, Feng J W, Zhu X T, et al. Evolution analyses of CAMTA transcription factor in plants and its enhancing effect on cold-tolerance. Frontiers in Plant Science, 2021, 12: 758187. |
| 4 | Kim Y, Gilmour S J, Chao L, et al. Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Molecular Plant, 2020, 13(1): 157-168. |
| 5 | Pei J, Flieder D B, Patchefsky A, et al. Detecting MYB and MYBL1 fusion genes in tracheobronchial adenoid cystic carcinoma by targeted RNA-sequencing. Modern Pathology, 2019, 32(10): 1416-1420. |
| 6 | Yang Y, Yoo C G, Rottmann W, et al. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biology, 2019, 19(1): 486. |
| 7 | Zhu D, Hou L X, Xiao P L, et al. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Science, 2019, 280: 132-142. |
| 8 | Lorenzo O. bZIP edgetic mutations: at the frontier of plant metabolism, development and stress trade-off. Journal of Experimental Botany, 2019, 70(20): 5517-5520. |
| 9 | Rahman H, Yang J, Xu Y P, et al. Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. oryzae. Frontiers in Plant Science, 2016, 7(303): 177-178. |
| 10 | Aravind L, Koonin E V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. Journal of Molecular Biology, 1999, 287(5): 1023-1040. |
| 11 | Müller C W, Rey F A, Sodeoka M, et al. Structure of the NF-κB p50 homodimer bound to DNA. Nature, 1995, 373(6512): 311-317. |
| 12 | Rubtsov A M, Lopina O D. Ankyrins. FEBS Letters, 2000, 482(1/2): 1-5. |
| 13 | Yang T, Poovaiah B W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. The Journal of Biological Chemistry, 2002, 277(47): 45049-45058. |
| 14 | Shkolnik D, Finkler A, Pasmanik-Chor M, et al. CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: a key regulator of Na+ homeostasis during germination. Plant Physiology, 2019, 180(2): 1101-1118. |
| 15 | Yuan P, Du L, Poovaiah B W. Ca2+/calmodulin-dependent AtSR1/CAMTA3 plays critical roles in balancing plant growth and immunity. International Journal of Molecular Sciences, 2018, 19(6): 1764-1765. |
| 16 | Satoshi K, Kazuo S, Kazuko Y. Transcriptional regulatory network of plant cold-stress responses. Trends in Plant Science, 2022, 27(9): 922-935. |
| 17 | Kim Y S, Park S C, Gilmour S J, et al. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. The Plant Journal: for Cell and Molecular Biology, 2013, 75(3): 364-376. |
| 18 | Yue R Q, Lu C X, Sun T, et al. Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Frontiers in Plant Science, 2015, 6(576): 576. |
| 19 | Yang T, Peng H, Whitaker B D, et al. Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiologia Plantarum, 2013, 148(3): 445-455. |
| 20 | Neha P, Alok R, Poonam P, et al. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics, 2013, 14(1): 216. |
| 21 | Yang F, Dong F S, Liu Y W, et al. Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) gene family in wheat (Triticum aestivum L.). BMC Genomics, 2020, 21(1): 105-106. |
| 22 | Leng X P, Han J, Wang X M, et al. Characterization of a calmodulin-binding transcription factor from strawberry (Fragaria×ananassa). The Plant Genome, 2015, 8(2): 1-12. |
| 23 | Liu Z P, Chen T L, Ma L C, et al. Global transcriptome sequencing using the Illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS One, 2013, 8(12): e83549. |
| 24 | Gain H, Nandi D, Kumari D S, et al. Genome-wide identification of CAMTA gene family members in rice (Oryza sativa L.) and in silico study on their versatility in respect to gene expression and promoter structure. Functional Integrative Genomics, 2022, 22(2): 193-214. |
| 25 | Shangguan L F, Wang X M, Leng X P, et al. Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera. Molecular Biology Reports, 2014, 41(5): 2937-2949. |
| 26 | Wang G P, Zeng H Q, Hu X Y, et al. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant and Soil, 2015, 386(1/2): 205-221. |
| 27 | Rahman H, Xu Y P, Zhang X R, et al. Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Frontiers in Plant Science, 2016, 7(177): 581. |
| 28 | Yang T, Peng H, Whitaker B D, et al. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biology, 2012, 12(1): 19-20. |
| 29 | Chen H, Zeng Y, Yang Y, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nature Communications, 2020, 11(1): 2494. |
| 30 | Yuan J, Amend A, Borkowski J, et al. MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics, 1999, 15(10): 862-863. |
| 31 | Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics, 1994, 10(2): 189-191. |
| 32 | Chen C, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 33 | Bailey T L, Boden M, Buske F A, et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Research, 2009, 37(2): W202-W208. |
| 34 | Luo D, Zhou Q, Wu Y, et al. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biology, 2019, 19(1): 1-20. |
| 35 | Luo D, Wu Y, Liu J, et al. Comparative transcriptomic and physiological analyses of Medicago sativa L. indicates that multiple regulatory networks are activated during continuous ABA treatment. International Journal of Molecular Sciences, 2018, 20(1): 1-22. |
| 36 | Min X, Liu Z, Wang Y, et al. Comparative transcriptomic analysis provides insights into the coordinated mechanisms of leaves and roots response to cold stress in common vetch. Industrial Crops Products, 2020, 158(1): 112949. |
| 37 | Luo Y, Liu Y B, Yu X D, et al. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment. Journal of Plant Physiology, 2009, 166(4): 385-394. |
| 38 | Ren M H, Zhang Y P, Xu T, et al. Identification and expression analyses of R2R3-MYB subfamily in alfalfa under drought stress. Acta Agrestia Sinica, 2023, 31(4): 972-983. |
| 任明辉, 张雨蓬, 许涛, 等. 紫花苜蓿R2R3-MYB亚家族鉴定与干旱胁迫下的表达分析. 草地学报, 2023, 31(4): 972-983. | |
| 39 | Yang Y J, Sun T, Xu L R, et al. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Frontiers in Plant Science, 2015, 6(459): 459. |
| 40 | Jiang X M, Yuan Z, Zhu F J, et al. Genome-wide identification and evolutionary analysis of CAMTA transcription factor family in Cenchrus fungigraminus. Journal of Fujian Agriculture and Forestry University (Natural Science Edition), 2023, 52(1): 10-16. |
| 姜晓梦, 袁振, 朱方捷, 等. 巨菌草(Cenchrus fungigraminus)全基因组CAMTA家族转录因子的鉴定及进化分析. 福建农林大学学报(自然科学版), 2023, 52(1): 10-16. | |
| 41 | Jiang H H, Li L, Li X F. Bioinformatics analysis of pepper CAMTA gene family. China Vegetables, 2022, 1(9): 47-56. |
| 蒋宏华, 李丽, 李雪峰. 辣椒CAMTA基因家族生物信息学分析. 中国蔬菜, 2022, 1(9): 47-56. | |
| 42 | Lin X Z, Xiao X H, Yang J H, et al. Genome-wide identification and expression analysis of the CAMTA family in rubber tree (Hevea brasiliensis). Chinese Journal of Tropical Crops, 2021, 42(10): 2859-2868. |
| 林显祖, 肖小虎, 阳江华, 等. 巴西橡胶树CAMTA转录因子全基因组鉴定与表达分析. 热带作物学报, 2021, 42(10): 2859-2868. |
| [1] | 刘昊, 李显炀, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 紫花苜蓿SAUR基因家族的鉴定及其在非生物胁迫中的表达模式研究[J]. 草业学报, 2024, 33(4): 135-153. |
| [2] | 李显炀, 刘昊, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 全基因组水平紫花苜蓿WRKY转录因子家族鉴定与表达模式分析[J]. 草业学报, 2024, 33(4): 154-170. |
| [3] | 李妍, 马富龙, 韩路, 王海珍. 美国‘WL’系列不同秋眠级苜蓿品种在南疆的生产性能与适应性评价[J]. 草业学报, 2024, 33(3): 139-149. |
| [4] | 孟超楠, 赵玉洁, 陈佳欣, 张旖璐, 王彦佳, 冯丽荣, 孙玉刚, 郭长虹. 2株青贮玉米根际固氮菌的筛选鉴定及促生作用研究[J]. 草业学报, 2024, 33(3): 174-185. |
| [5] | 汪雪, 刘晓静, 王静, 吴勇, 童长春. 连续间作下的紫花苜蓿/燕麦根系与碳氮代谢特性研究[J]. 草业学报, 2024, 33(3): 85-96. |
| [6] | 唐璎, 刘晓静, 赵雅姣, 董霖. 甘肃不同区域青贮紫花苜蓿乳酸菌群落特征及其驱动因子研究[J]. 草业学报, 2024, 33(2): 112-124. |
| [7] | 陈嘉慧, 刘文献. 重要牧草组学数据图形可视化展示工具的构建及应用[J]. 草业学报, 2024, 33(2): 57-67. |
| [8] | 白旭琴, 贾春云, 李文栓, 李亚敏, 刘长风, 韩秀云, 褚美函, 巩宗强, 李晓军. 叶面喷施硒肥对紫花苜蓿富硒降镉效果的影响[J]. 草业学报, 2024, 33(1): 50-60. |
| [9] | 刘选帅, 孙延亮, 马春晖, 张前兵. 菌磷耦合下紫花苜蓿的干物质产量及磷素空间分布特征[J]. 草业学报, 2023, 32(9): 104-115. |
| [10] | 徐蕊, 王峥, 王仪明, 苏连泰, 高鲤, 周鹏, 安渊. 紫花苜蓿对轮作水稻产量和蔗糖代谢的影响[J]. 草业学报, 2023, 32(8): 129-140. |
| [11] | 王宝强, 马文静, 王贤, 朱晓林, 赵颖, 魏小红. 一氧化氮对干旱胁迫下紫花苜蓿幼苗次生代谢产物的影响[J]. 草业学报, 2023, 32(8): 141-151. |
| [12] | 凌文卿, 张磊, 李珏, 冯启贤, 李妍, 周燚, 刘一佳, 阳伏林, 周晶. 布氏乳杆菌和不同糖类联用对紫花苜蓿青贮营养成分、发酵品质、瘤胃降解率及有氧稳定性的影响[J]. 草业学报, 2023, 32(7): 122-134. |
| [13] | 王少鹏, 刘佳, 洪军, 林积圳, 张义, 史昆, 王赞. 紫花苜蓿MsPPR1基因的克隆及抗旱功能分析[J]. 草业学报, 2023, 32(7): 49-60. |
| [14] | 管瑾, 郭一荻, 刘凌云, 尹淑霞, 滕珂. 结缕草叶肉细胞原生质体瞬时基因表达系统的构建[J]. 草业学报, 2023, 32(7): 61-71. |
| [15] | 张振粉, 黄荣, 李向阳, 姚博, 赵桂琴. 基于Illumina MiSeq高通量测序的燕麦种带细菌多样性及功能分析[J]. 草业学报, 2023, 32(7): 96-108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||