

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2025, Vol. 34 ›› Issue (12): 157-169.DOI: 10.11686/cyxb2025014
• 研究论文 • 上一篇
李永龙( ), 周生辉, 薛梦瑶, 高远, 巨乐, 陈奕冰, 付松林, 郝建昊, 李恒, 张昆(
), 周生辉, 薛梦瑶, 高远, 巨乐, 陈奕冰, 付松林, 郝建昊, 李恒, 张昆( ), 左志芳(
), 左志芳( )
)
收稿日期:2025-01-17
修回日期:2025-03-27
出版日期:2025-12-20
发布日期:2025-10-20
通讯作者:
张昆,左志芳
作者简介:E-mail: zk61603@163.com基金资助:
Yong-long LI( ), Sheng-hui ZHOU, Meng-yao XUE, Yuan GAO, Le JU, Yi-bing CHEN, Song-lin FU, Jian-hao HAO, Heng LI, Kun ZHANG(
), Sheng-hui ZHOU, Meng-yao XUE, Yuan GAO, Le JU, Yi-bing CHEN, Song-lin FU, Jian-hao HAO, Heng LI, Kun ZHANG( ), Zhi-fang ZUO(
), Zhi-fang ZUO( )
)
Received:2025-01-17
Revised:2025-03-27
Online:2025-12-20
Published:2025-10-20
Contact:
Kun ZHANG,Zhi-fang ZUO
摘要:
WRKY转录因子在植物生长发育、病原菌防御和非生物胁迫应答过程中发挥重要调控作用。根据前期结缕草中WRKY转录因子的基因家族鉴定以及耐盐和敏盐结缕草品系在盐胁迫下的转录组分析,筛选得到1个盐胁迫响应WRKY转录因子基因ZjWRKY63。为进一步研究该基因的耐盐性功能,设计引物进行PCR扩增,克隆得到开放阅读框为921 bp的基因编码区(CDS)序列。ZjWRKY63基因编码1个含有306个氨基酸的蛋白质,具有1个WRKY功能保守结构域和1个C2H2锌指结构基序,属于WRKY IIa亚族。生物信息学分析表明,ZjWRKY63氨基酸残基平均信号肽最大得分值为0.267,提示该蛋白不具备信号肽;疏水性预测分析显示编码蛋白总平均疏水指数(GRAVY)为-0.395,提示该蛋白质为亲水性蛋白质,且ZjWRKY63不存在跨膜区;亚细胞定位预测分析显示该转录因子定位于细胞核内。系统发育分析显示,结缕草ZjWRKY63与芦苇 PaWRKY65亲缘关系最近。对过表达ZjWRKY63的转基因拟南芥种子和植株分别进行盐胁迫处理发现,过表达植株的种子萌发率、存活率和侧根数均显著高于野生型。同时,在ZjWRKY63转基因拟南芥中,盐胁迫响应相关基因的表达量均显著高于野生型,推测ZjWRKY63基因通过调节胁迫响应基因的表达来提高转基因拟南芥的耐盐性。以上研究结果初步揭示了结缕草ZjWRKY63基因的耐盐性功能,为深入研究ZjWRKY63基因的耐盐分子机制奠定了理论基础。
李永龙, 周生辉, 薛梦瑶, 高远, 巨乐, 陈奕冰, 付松林, 郝建昊, 李恒, 张昆, 左志芳. 结缕草ZjWRKY63基因的克隆及转基因拟南芥的耐盐性分析[J]. 草业学报, 2025, 34(12): 157-169.
Yong-long LI, Sheng-hui ZHOU, Meng-yao XUE, Yuan GAO, Le JU, Yi-bing CHEN, Song-lin FU, Jian-hao HAO, Heng LI, Kun ZHANG, Zhi-fang ZUO. Cloning of the gene ZjWRKY63 from Zoysia japonica and its salt resistance analysis in transgenic Arabidopsis[J]. Acta Prataculturae Sinica, 2025, 34(12): 157-169.
| 名称Name | 寡核苷酸Oligonucleotides (5′-3′) | 用途Purpose |
|---|---|---|
| ZjWRKY63-F | GTGTTACTTCCCCGGGGATCCATGGACAGCGAGTGGAGCG | ZjWRKY63基因克隆 For the gene cloning of ZjWRKY63 |
| ZjWRKY63-R | GTCATCCTTGTAGTCAAGCTTTGTCATGGCAGTGCCTCC | |
| AtABI5-F | AGAGGGATAGCGAACGAGTCTAGTC | 用于下游基因的qRT-PCR表达分析 For the qRT-PCR of downstream gene expression analysis |
| AtABI5-R | GTTCGGGTTTGGATTAGGTTTAGG | |
| AtCOR15A-F | CAGTTCGTCGTCGTTTCT | |
| AtCOR15A-R | CCAATGTATCTGCGGTTT | |
| AtDREB1A-F | AGGAGACGTTGGTGGAGGCT | |
| AtDREB1A-R | ACGTCGTCATCATCGCCGTC | |
| AtMYC2-F | GCGTTGATGGATTTGGAGTT | |
| AtMYC2-R | TTGCTCTGAGCTGTTCTTGC | |
| AtSOS1-F | TCGTTTCAGCCAAATCAGAAAGT | |
| AtSOS1-R | TTTGCCTTGTGCTGCTTTCC | |
| AtSOS2-F | GGCTTGAAGAAAGTGAGTCTCG | |
| AtSOS2-R | GCTACATAGTTCGGAGTTCCACA | |
| AtACT-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参基因 Inner reference gene of A. thaliana |
| AtACT-R | AACGACCTTAATCTTCATGCTGC |
表1 基因克隆和表达分析所用引物
Table 1 Primers used for gene cloning and expression analysis
| 名称Name | 寡核苷酸Oligonucleotides (5′-3′) | 用途Purpose |
|---|---|---|
| ZjWRKY63-F | GTGTTACTTCCCCGGGGATCCATGGACAGCGAGTGGAGCG | ZjWRKY63基因克隆 For the gene cloning of ZjWRKY63 |
| ZjWRKY63-R | GTCATCCTTGTAGTCAAGCTTTGTCATGGCAGTGCCTCC | |
| AtABI5-F | AGAGGGATAGCGAACGAGTCTAGTC | 用于下游基因的qRT-PCR表达分析 For the qRT-PCR of downstream gene expression analysis |
| AtABI5-R | GTTCGGGTTTGGATTAGGTTTAGG | |
| AtCOR15A-F | CAGTTCGTCGTCGTTTCT | |
| AtCOR15A-R | CCAATGTATCTGCGGTTT | |
| AtDREB1A-F | AGGAGACGTTGGTGGAGGCT | |
| AtDREB1A-R | ACGTCGTCATCATCGCCGTC | |
| AtMYC2-F | GCGTTGATGGATTTGGAGTT | |
| AtMYC2-R | TTGCTCTGAGCTGTTCTTGC | |
| AtSOS1-F | TCGTTTCAGCCAAATCAGAAAGT | |
| AtSOS1-R | TTTGCCTTGTGCTGCTTTCC | |
| AtSOS2-F | GGCTTGAAGAAAGTGAGTCTCG | |
| AtSOS2-R | GCTACATAGTTCGGAGTTCCACA | |
| AtACT-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参基因 Inner reference gene of A. thaliana |
| AtACT-R | AACGACCTTAATCTTCATGCTGC |

图1 结缕草ZjWRKY63基因的转录组及克隆分析A: 盐胁迫下ZjWRKY差异表达基因在结缕草耐盐材料Z011和敏盐材料Z004中的转录组分析Transcriptome analysis of differentially expressed genes of ZjWRKY in the salt-tolerant material Z011 and the salt-sensitive material Z004 of Z. japonica under salt stress; B: ZjWRKY63基因在Z011和Z004中的转录组分析Transcriptome analysis of ZjWRKY63 in Z011 and Z004; C: ZjWRKY63基因的克隆 Cloning of ZjWRKY63. FPKM: 每百万个读数来估算的基因表达量Fragments per kilobase million; M: Marker.
Fig.1 Transcriptome and cloning analysis of Z. japonica ZjWRKY63

图2 结缕草ZjWRKY63保守结构域分析A: ZjWRKY63保守结构域预测Prediction of ZjWRKY63 conserved domains; B: ZjWRKY63基因的核酸序列及其编码的氨基酸序列 Nucleotide sequence and encoded amino acid sequence of ZjWRKY63. 虚线为WRKYGQK保守基序 The dotted line indicates the conserved motif of WRKYGQK; 实线为C2H2基序(C-X5-C-X23-H-X-H) The solid line represents the C2H2 motif of C-X5-C-X23-H-X-H.
Fig.2 The conserved domain analysis of Z. japonicaZjWRKY63
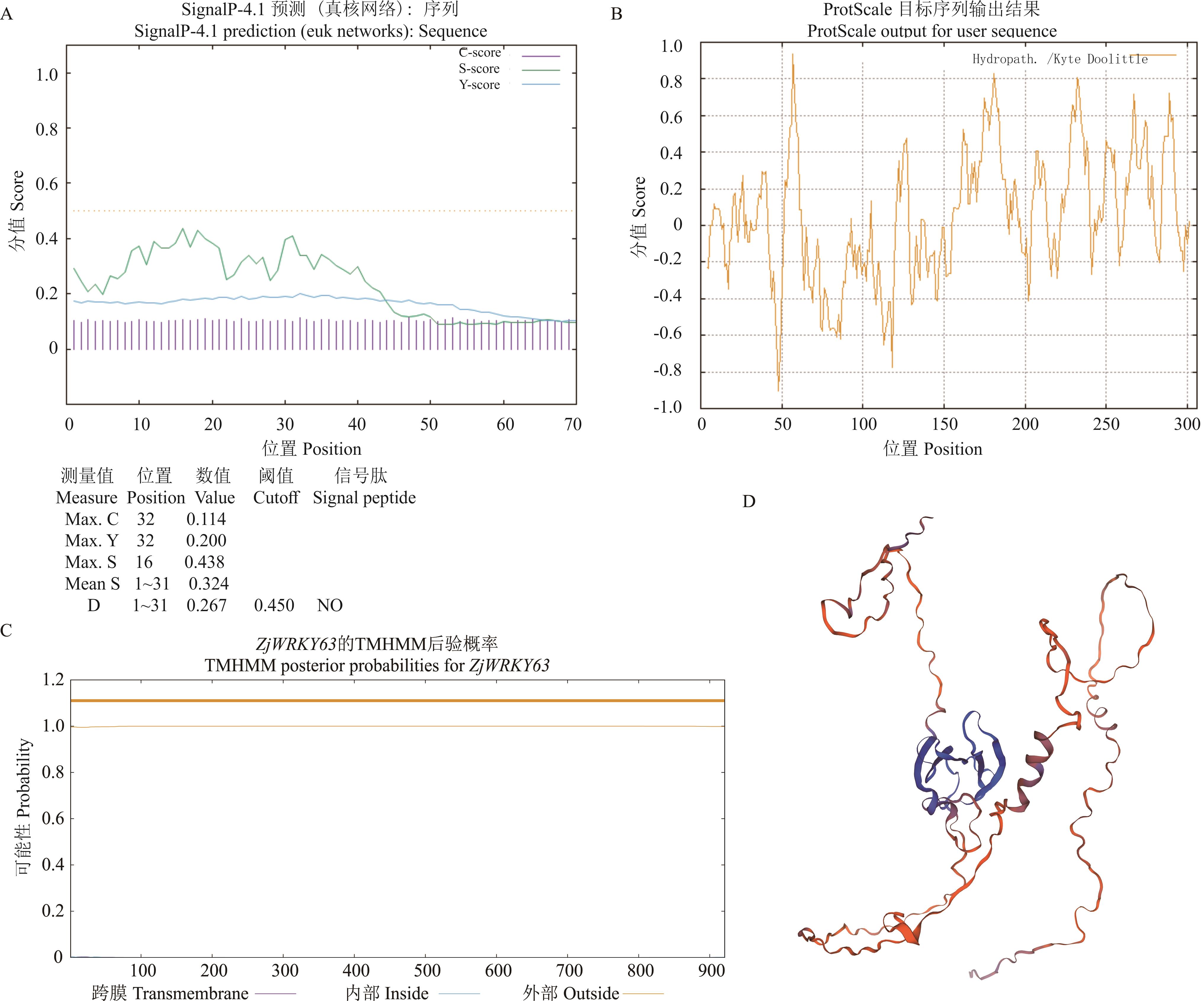
图3 结缕草ZjWRKY63基因生物信息学分析A: ZjWRKY63蛋白信号肽预测 Prediction of ZjWRKY63 signal peptide; B: ZjWRKY63蛋白疏水性预测Prediction of ZjWRKY63 hydrophobic property; C: ZjWRKY63蛋白跨膜结构预测 Prediction of ZjWRKY63 transmembrane structure; D: ZjWRKY63蛋白三级结构预测 Prediction of ZjWRKY63 protein tertiary structure.
Fig.3 Bioinformatic analysis of Z. japonica ZjWRKY63
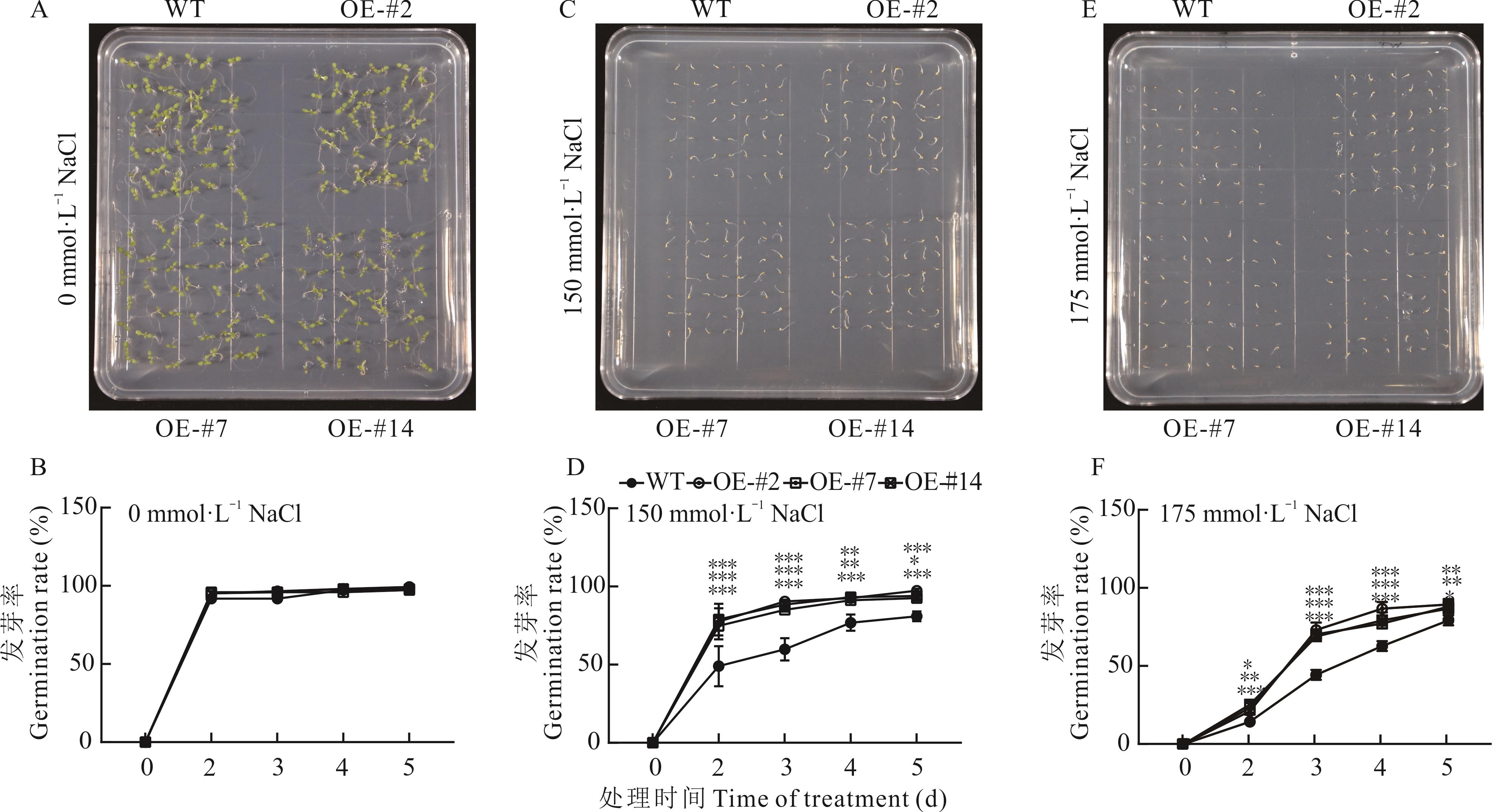
图5 过表达ZjWRKY63转基因拟南芥种子萌发率分析A, B: 0 mmol·L-1 NaCl 处理后的种子萌发率 The germination rate under 0 mmol·L-1 NaCl treatment; C, D: 150 mmol·L-1 NaCl 处理后的种子萌发率 The germination rate under 150 mmol·L-1 NaCl treatment; E, F: 175 mmol·L-1 NaCl 处理后的种子萌发率 The germination rate under 175 mmol·L-1 NaCl treatment; *表示P<0. 05 * represents P<0.05; **表示P<0.01 ** represents P<0.01; ***表示P<0.001 *** represents P<0.001;下同The same below.
Fig.5 Germination rate analysis of transgenic A. thaliana with overexpression of ZjWRKY63
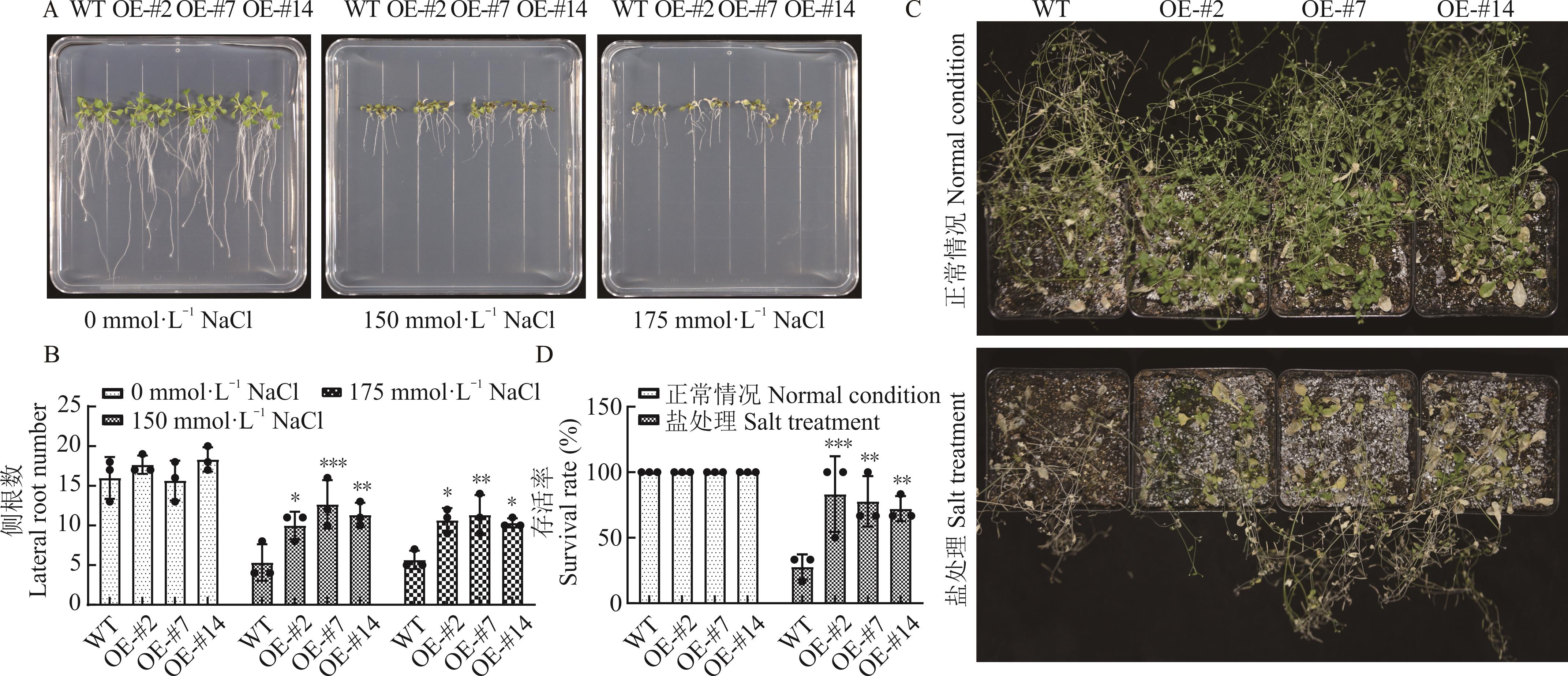
图6 过表达ZjWRKY63转基因拟南芥耐盐性分析A, B: 不同浓度NaCl处理下转基因各株系和野生型的侧根数The lateral root number of transgenic plants and wild type under different NaCl treatments; C, D: 盐胁迫处理后转基因各株系和野生型的存活率The survival rate of transgenic plants and wild type after salt stress treatment.
Fig.6 The salt tolerant analysis of transgenic A. thaliana with overexpression of ZjWRKY63
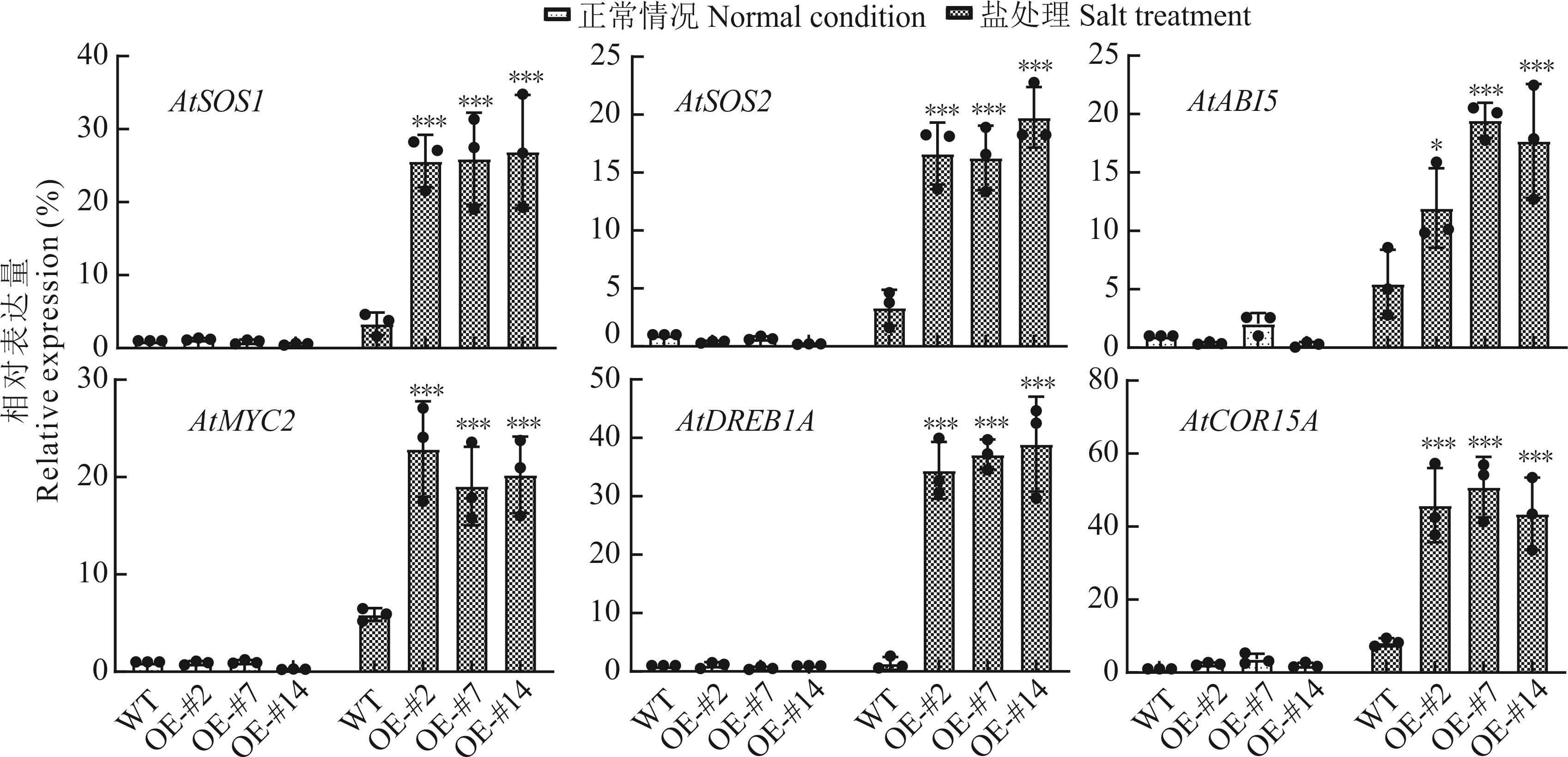
图7 盐胁迫处理下ZjWRKY63转基因拟南芥中胁迫响应基因的表达分析
Fig.7 The expression analysis of stress-responsive genes in transgenic A. thaliana with overexpression of ZjWRKY63 under salt stress treatment
| [1] | Liu Y, Jiang W, Zhao W, et al. Effects of biochar application on soil properties and the growth of Melissa officinalis L. under salt stress. Scientia Horticulturae, 2024, 338(1): 113704. |
| [2] | Li J X, Bai X M, Zhang C, et al. Effects of different salt types on seed germination and seedling growth of Poa annua. Arid Zone Research, 2023, 40(7): 1131-1140. |
| 李娟霞, 白小明, 张翠, 等. 不同盐分类型对一年生早熟禾种子萌发和幼苗生长的影响. 干旱区研究, 2023, 40(7): 1131-1140. | |
| [3] | Wang R, Yu M Y, Cao Z J, et al. Effects of salt stress on seed germination characteristics of Poa pratensis L. Seed, 2024, 43(9): 86-93, 100. |
| 王蓉, 于铭玥, 曹志坚, 等. 盐处理对草地早熟禾种子萌发特性的影响. 种子, 2024, 43(9): 86-93, 100. | |
| [4] | Song X. Research progress on stress resistance of seashore paspalum. Modern Agricultural Science and Technology, 2024(11): 126-132. |
| 宋鑫. 海滨雀稗抗逆性研究进展. 现代农业科技, 2024(11): 126-132. | |
| [5] | Zuo Z F, Lee H Y, Kang H G. Basic helix-loop-helix transcription factors: regulators for plant growth development and abiotic stress responses. International Journal of Molecular Sciences, 2023, 24(2): 1419. |
| [6] | Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. Journal of Experimental Botany, 2011, 62(14): 4731-4748. |
| [7] | Rai G K, Mishra S, Chouhan R, et al. Plant salinity stress, sensing, and its mitigation through WRKY. Frontiers in Plant Science, 2023, 14(4): 1238507. |
| [8] | Johnson C S, Kolevski B, Smyth D R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell, 2002, 14(6): 1359-1375. |
| [9] | Rushton P J, Somssich I E, Ringler P, et al. WRKY transcription factors. Trends in Plant Science, 2010, 15(5): 247-258. |
| [10] | Eulgem T, Rushton P J, Robatzek S, et al. The WRKY superfamily of plant transcription factors. Trends in Plant Science, 2000, 5(5): 199-206. |
| [11] | Dong J X, Chen C H, Chen Z X, et al. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology, 2003, 51(1): 21-37. |
| [12] | Wu K L, Guo Z J, Wang H H, et al. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research, 2005, 12(1): 9-26. |
| [13] | Eulgem T, Somssich I E. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology, 2007, 10(4): 366-371. |
| [14] | Jiang J J, Ma S H, Ye N H, et al. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 2017, 59(2): 86-101. |
| [15] | Liang C P, Chen L G. Preliminary exploration of the mechanism by which AtWRKY45 regulates salt tolerance in Arabidopsis thaliana. Seed Science and Technology, 2024, 42(17): 13-17. |
| 梁承萍, 陈利钢. AtWRKY45调控拟南芥耐盐性的机制初探. 种子科技, 2024, 42(17): 13-17. | |
| [16] | Zhou S, Zheng W J, Liu B H, et al. Characterizing the role of TaWRKY13 in salt tolerance. International Journal of Molecular Sciences, 2019, 20(22): 5712. |
| [17] | Zhu D, Hou L X, Xiao P L, et al. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Science, 2018, 280: 132-142. |
| [18] | Li X, Ye G, Shen Z, et al. Na+ and K+ homeostasis in different organs of contrasting Zoysia japonica accessions under salt stress. Environmental and Experimental Botany, 2023, 214: 105455. |
| [19] | Bao W, Wang X, Chen M, et al. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Reports, 2018, 37(7): 1033-1048. |
| [20] | Cai R, Dai W, Zhang C, et al. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta, 2017, 246: 1215-1231. |
| [21] | Zhao K, Zhang D, Lv K, et al. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Science, 2019, 289: 110259. |
| [22] | Yuan Y, Son J H, Park M Y, et al. Overexpression of ZjWRKY10, a Zoysia japonica WRKY transcription factor gene, accelerates leaf senescence and flowering in transgenic Arabidopsis. Journal of Plant Biotechnology, 2024, 51(1): 1-10. |
| [23] | He X, Duan H L, Luo L J, et al. Morphological characteristics variations of germplasm resources in Zoysia Willd. Molecular Plant Breeding, 2024, 22(13): 4364-4376. |
| 何潇, 段宏利, 罗丽娟, 等. 结缕草属种质资源形态特征变异. 分子植物育种, 2024, 22(13): 4364-4376. | |
| [24] | Li H, Li D F, Deng Y, et al. Expression analysis of abiotic stress response gene HcWRKY71 in kenaf and transformation of Arabidopsis. Acta Agronomica Sinica, 2021, 47(6): 1090-1099. |
| 李辉, 李德芳, 邓勇, 等. 红麻非生物逆境胁迫响应基因HCWRKY71表达分析及转化拟南芥. 作物学报, 2021, 47(6): 1090-1099. | |
| [25] | Li Y, Mi X F, Yan L, et al. Cloning and salt tolerance analysis of UDP-glycosyltransferases UGT72B3 gene in Carex rigescens. Chinese Journal of Grassland, 2023, 45(8): 10-22. |
| 李岩, 米鑫丰, 闫丽, 等. 白颖苔草UDP-糖基转移酶UGT72B3基因的克隆与耐盐性分析. 中国草地学报, 2023, 45(8): 10-22. | |
| [26] | Li Y, Yang Q, Huang H, et al. Overexpression of PvWAK3 from seashore paspalum increases salt tolerance in transgenic Arabidopsis via maintenance of ion and ROS homeostasis. Plant Physiology and Biochemistry, 2024, 207: 108337. |
| [27] | Yu Y G, Wu Y X, He L. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Molecular Biology, 2023, 113(4): 171-191. |
| [28] | Wang J J, An C, Guo H L, et al. Physiological and transcriptomic analyses reveal the mechanisms underlying the salt tolerance of Zoysia japonica Steud. BMC Plant Biology, 2020, 20: 1-16. |
| [29] | Yamasaki K, Kigawa T, Seki M, et al. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends in Plant Science, 2013, 18(5): 267-276. |
| [30] | Zhang H Y, Liu Y W, Yang J F, et al. Identification and analysis of salt tolerance of wheat transcription factor TaWRKY33 protein. Scientia Agricultura Sinica, 2018, 51(24): 4591-4602. |
| 张惠媛, 刘永伟, 杨军峰, 等. 小麦转录因子基因TaWRKY33的耐盐性分析. 中国农业科学, 2018, 51(24): 4591-4602. | |
| [31] | Qin Y, Tian Y, Liu X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochemical and Biophysical Research Communications, 2015, 464(2): 428-433. |
| [32] | Lv B B, Wu Q, Wang A H, et al. A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiology and Biochemistry, 2020, 147: 43-53. |
| [33] | Li P L, Song A P, Gao C Y, et al. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Reports, 2015, 34(8): 1365-1378. |
| [34] | Muchate N S, Nikalje G C, Rajurkar N S, et al. Plant salt stress: adaptive responses, tolerance mechanism and bioengineering for salt tolerance. The Botanical Review, 2016, 82(4): 371-406. |
| [35] | Wang Q, Guan C, Wang P, et al. The effect of AtHKT1; 1 or AtSOS1 mutation on the expressions of Na+ or K+ transporter genes and ion homeostasis in Arabidopsis thaliana under salt stress. International Journal of Molecular Sciences, 2019, 20(5): 1085. |
| [36] | Shi H Z, Zhu J K. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Molecular Biology, 2002, 50(3): 543-550. |
| [37] | Ryu H, Cho Y G. Plant hormones in salt stress tolerance. Journal of Plant Biology, 2015, 58(3): 147-155. |
| [38] | Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Archives of Biochemistry and Biophysics, 2005, 444(2): 139-158. |
| [1] | 邹苇鹏, 刘怡, 翟佳兴, 周思懿, 宫祉祎, 岑慧芳, 朱慧森, 许涛. 紫花苜蓿MsNAC053基因克隆及其对非生物胁迫的响应分析[J]. 草业学报, 2025, 34(9): 121-133. |
| [2] | 赵媛媛, 蒲小剑, 徐成体, 王伟, 傅云洁. 蒺藜苜蓿MtBMI1基因克隆及抗旱性分析[J]. 草业学报, 2025, 34(6): 139-153. |
| [3] | 左志芳, 李永龙, 魏雨佳, 周生辉, 李岩, 杨国锋. 结缕草DREB基因家族的鉴定及非生物胁迫下的表达模式分析[J]. 草业学报, 2025, 34(5): 74-88. |
| [4] | 周昕越, 王丽萍, 蒋庆雪, 马晓冉, 仪登霞, 王学敏. 紫花苜蓿低温诱导蛋白MsLTI65的分离及其对不同逆境的响应[J]. 草业学报, 2025, 34(5): 89-104. |
| [5] | 林心怡, 王旎, 陈拓, 宋一岚, 陆耀东, 董朝霞. 3种植物生长调节剂对结缕草耐荫性的影响[J]. 草业学报, 2025, 34(3): 224-232. |
| [6] | 汪欣瑶, 彭亚萍, 姚立蓉, 汪军成, 司二静, 张宏, 杨轲, 马小乐, 孟亚雄, 王化俊, 李葆春. 盐生草HgS5基因的克隆与抗旱性鉴定[J]. 草业学报, 2025, 34(2): 184-195. |
| [7] | 孟晨, 鲁雪莉, 宋亦汝, 张成省, 李义强, 项海芹, 徐宗昌. 11份益母草种质材料苗期耐盐性评价与鉴定[J]. 草业学报, 2024, 33(5): 196-203. |
| [8] | 王萌, 鲁雪莉, 王菊英, 张梦超, 宋奕汝, 孟晨, 张莉, 徐宗昌. 小黑麦种质萌发期苗期耐盐资源评价与筛选[J]. 草业学报, 2024, 33(5): 58-68. |
| [9] | 张译尹, 李雪颖, 王斌, 宋珂辰, 兰剑, 胡海英. 盐胁迫对不同种质小黑麦幼苗水分利用效率和渗透调节的影响[J]. 草业学报, 2024, 33(4): 87-98. |
| [10] | 周昕越, 蒋庆雪, 贾会丽, 马琳, 樊璐, 王学敏. 紫花苜蓿MsBBX20基因克隆及耐盐功能分析[J]. 草业学报, 2024, 33(10): 55-73. |
| [11] | 胡尚钦, 汪军成, 姚立蓉, 司二静, 马小乐, 杨轲, 张宏, 孟亚雄, 王化俊, 李葆春. 盐生草根系基因HgAKR6C的克隆与初步功能分析[J]. 草业学报, 2024, 33(1): 61-74. |
| [12] | 管瑾, 郭一荻, 刘凌云, 尹淑霞, 滕珂. 结缕草叶肉细胞原生质体瞬时基因表达系统的构建[J]. 草业学报, 2023, 32(7): 61-71. |
| [13] | 杨小涵, 伍国强, 魏明, 王北辰. HKT在植物离子稳态和响应非生物逆境胁迫中的作用[J]. 草业学报, 2023, 32(5): 190-202. |
| [14] | 王园, 王晶, 李淑霞. 紫花苜蓿MsBBX24基因的克隆及耐盐性分析[J]. 草业学报, 2023, 32(3): 107-117. |
| [15] | 苗涵, 魏莱, 杨燕萍, 车永和. 海水胁迫下冰草幼苗期耐盐性指标筛选[J]. 草业学报, 2023, 32(3): 200-211. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||