

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2025, Vol. 34 ›› Issue (4): 124-136.DOI: 10.11686/cyxb2024206
• 研究论文 • 上一篇
罗天蓉1( ), 马健芝1, 杜明阳1, 多杰措2, 熊辉岩2, 段瑞君1(
), 马健芝1, 杜明阳1, 多杰措2, 熊辉岩2, 段瑞君1( )
)
收稿日期:2024-06-03
修回日期:2024-07-22
出版日期:2025-04-20
发布日期:2025-02-19
通讯作者:
段瑞君
作者简介:Corresponding author. E-mail: ruijunduan@163.com基金资助:
Tian-rong LUO1( ), Jian-zhi MA1, Ming-yang DU1, Jie-cuo DUO2, Hui-yan XIONG2, Rui-jun DUAN1(
), Jian-zhi MA1, Ming-yang DU1, Jie-cuo DUO2, Hui-yan XIONG2, Rui-jun DUAN1( )
)
Received:2024-06-03
Revised:2024-07-22
Online:2025-04-20
Published:2025-02-19
Contact:
Rui-jun DUAN
摘要:
长链脂酰辅酶A合成酶(LACS)基因家族是包含在酰基激活酶超家族中的一类,在脂肪酸合成代谢中具有重要的作用。基于紫花苜蓿基因组数据,采用生物信息学方法对紫花苜蓿LACS基因家族成员进行鉴定,构建系统发育树,进行理化性质、染色体定位、保守基序和基因结构、顺式作用原件及组织特异性表达分析,然后构建蛋白互作网络分析,通过qRT-PCR试验进行非生物胁迫表达分析。结果表明:在紫花苜蓿基因组中共鉴定得到10个MsLACS家族成员,分别位于5条染色体上;通过构建系统发育树,将MsLACS分为5支;保守基序分析发现12个不同的保守基序,Motif3构成了AMP保守结构域,MsLACS基因均含10~12个基序不等;基因结构分析发现,MsLACS基因结构大有不同,MsLACS基因的外显子数量为11~22个,所含内含子数量为0~3个不等,MsLACS1-1和MsLACS3不含内含子;MsLACS基因的启动子区域中主要有光反应元件、激素反应元件和非生物胁迫反应元件等;MsLACS基因在不同组织中表达不同,且具有明显的组织特异性;非生物胁迫表达分析表明:紫花苜蓿MsLACS基因对于干旱胁迫和盐胁迫响应水平整体较高,对冷胁迫响应水平偏低,表达量整体呈先升高后降低的起伏型,在叶片组织中偏高,根组织中较低;干旱和盐胁迫下,MsLACS基因在叶片中均高表达,胁迫时间段为6 h时,表达量水平整体达到最高;紫花苜蓿10个MsLACS基因共同互作,各蛋白与其他蛋白互作连线分别有18条,相互间有较强互作。研究结果可为探究紫花苜蓿中LACS基因表达以及胁迫育种提供一定研究基础。
罗天蓉, 马健芝, 杜明阳, 多杰措, 熊辉岩, 段瑞君. 紫花苜蓿LACS基因家族成员鉴定及表达分析[J]. 草业学报, 2025, 34(4): 124-136.
Tian-rong LUO, Jian-zhi MA, Ming-yang DU, Jie-cuo DUO, Hui-yan XIONG, Rui-jun DUAN. Identification and expression analysis of LACS gene family members in Medicago sativa[J]. Acta Prataculturae Sinica, 2025, 34(4): 124-136.
| 基因Gene | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (3′-5′) |
|---|---|---|
| MsLACS1-1 | CATGCTGGGATGGCGTAAAAT | AGTGAAGCACTTTTTGCAACC |
| MsLACS1-2 | TACCATGGGGTCAAAGCTCG | CCAACAGTACCAAGCATGCAC |
| MsLACS2 | TTGGTTTCATACAGCCATGAAAAT | TTCTGTCGGGGACTACCACA |
| MsLACS3 | CGGGGCTGTGGAGTTTGTTA | CATGACCCCCGATCTTCTCA |
| MsLACS5 | GGAACGGGACCTTATCACTCC | GCAGTCAGTAAAAGACAGCTCAT |
| MsLACS8 | ACTGGTGACATTGGGCGATT | CCAGAGACTGACGTGAAGCA |
| MsLACS9-2 | TTCAGGGTTGTTTCAGGCGA | CAGCCGTGTCCAACAGATGA |
| Msactin | CAAAAGATGGCAGATGCTGAGGAT | CATGACACCAGTATGACGAGGTCG |
表1 qRT-PCR引物
Table 1 qRT-PCR primers
| 基因Gene | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (3′-5′) |
|---|---|---|
| MsLACS1-1 | CATGCTGGGATGGCGTAAAAT | AGTGAAGCACTTTTTGCAACC |
| MsLACS1-2 | TACCATGGGGTCAAAGCTCG | CCAACAGTACCAAGCATGCAC |
| MsLACS2 | TTGGTTTCATACAGCCATGAAAAT | TTCTGTCGGGGACTACCACA |
| MsLACS3 | CGGGGCTGTGGAGTTTGTTA | CATGACCCCCGATCTTCTCA |
| MsLACS5 | GGAACGGGACCTTATCACTCC | GCAGTCAGTAAAAGACAGCTCAT |
| MsLACS8 | ACTGGTGACATTGGGCGATT | CCAGAGACTGACGTGAAGCA |
| MsLACS9-2 | TTCAGGGTTGTTTCAGGCGA | CAGCCGTGTCCAACAGATGA |
| Msactin | CAAAAGATGGCAGATGCTGAGGAT | CATGACACCAGTATGACGAGGTCG |
基因 Gene | 基因ID号 Gene ID | 氨基酸数 Number of amino acids (aa) | 分子质量 Molecular mass (kD) | 等电 点pI | 不稳定系数 Instability coefficient | 脂溶性系数 Fat solubility coefficient | 染色体定位 Chromosomal localization | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| MsLACS1-1 | MsG0180004774.01.T01 | 610 | 27.09 | 88.48 | Chr1 | 叶绿体Chloroplast | ||
| MsLACS1-2 | MsG0780041383.01.T01 | 597 | 31.70 | 84.00 | Chr7 | 细胞质Cytoplasm | ||
| MsLACS2 | MsG0180001276.01.T02 | 654 | 73203.98 | 5.74 | 40.29 | 86.90 | Chr1 | 细胞核Cell nucleus |
| MsLACS3 | MsG0480023684.01.T01 | 584 | Chr4 | 细胞质Cytoplasm | ||||
| MsLACS4 | MsG0580024346.01.T01 | 662 | 6.37 | 33.60 | 89.18 | Chr5 | 细胞核Cell nucleus | |
| MsLACS5 | MsG0580024344.01.T01 | 744 | Chr5 | 细胞质Cytoplasm | ||||
| MsLACS6 | MsG0180005634.01.T01 | 692 | 6.35 | 29.23 | 88.03 | Chr1 | 细胞质Cytoplasm | |
| MsLACS8 | MsG0380016055.01.T02 | 727 | 7.19 | 29.26 | 93.59 | Chr3 | 叶绿体Chloroplast | |
| MsLACS9-1 | MsG0380016352.01.T01 | 860 | Chr3 | 叶绿体Chloroplast | ||||
| MsLACS9-2 | MsG0180001085.01.T01 | 697 | 6.51 | 30.95 | 95.24 | Chr1 | 叶绿体Chloroplast |
表2 MsLACS基因编码蛋白理化性质分析
Table 2 Physicochemical property analysis of MsLACS gene encoded protein
基因 Gene | 基因ID号 Gene ID | 氨基酸数 Number of amino acids (aa) | 分子质量 Molecular mass (kD) | 等电 点pI | 不稳定系数 Instability coefficient | 脂溶性系数 Fat solubility coefficient | 染色体定位 Chromosomal localization | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| MsLACS1-1 | MsG0180004774.01.T01 | 610 | 27.09 | 88.48 | Chr1 | 叶绿体Chloroplast | ||
| MsLACS1-2 | MsG0780041383.01.T01 | 597 | 31.70 | 84.00 | Chr7 | 细胞质Cytoplasm | ||
| MsLACS2 | MsG0180001276.01.T02 | 654 | 73203.98 | 5.74 | 40.29 | 86.90 | Chr1 | 细胞核Cell nucleus |
| MsLACS3 | MsG0480023684.01.T01 | 584 | Chr4 | 细胞质Cytoplasm | ||||
| MsLACS4 | MsG0580024346.01.T01 | 662 | 6.37 | 33.60 | 89.18 | Chr5 | 细胞核Cell nucleus | |
| MsLACS5 | MsG0580024344.01.T01 | 744 | Chr5 | 细胞质Cytoplasm | ||||
| MsLACS6 | MsG0180005634.01.T01 | 692 | 6.35 | 29.23 | 88.03 | Chr1 | 细胞质Cytoplasm | |
| MsLACS8 | MsG0380016055.01.T02 | 727 | 7.19 | 29.26 | 93.59 | Chr3 | 叶绿体Chloroplast | |
| MsLACS9-1 | MsG0380016352.01.T01 | 860 | Chr3 | 叶绿体Chloroplast | ||||
| MsLACS9-2 | MsG0180001085.01.T01 | 697 | 6.51 | 30.95 | 95.24 | Chr1 | 叶绿体Chloroplast |
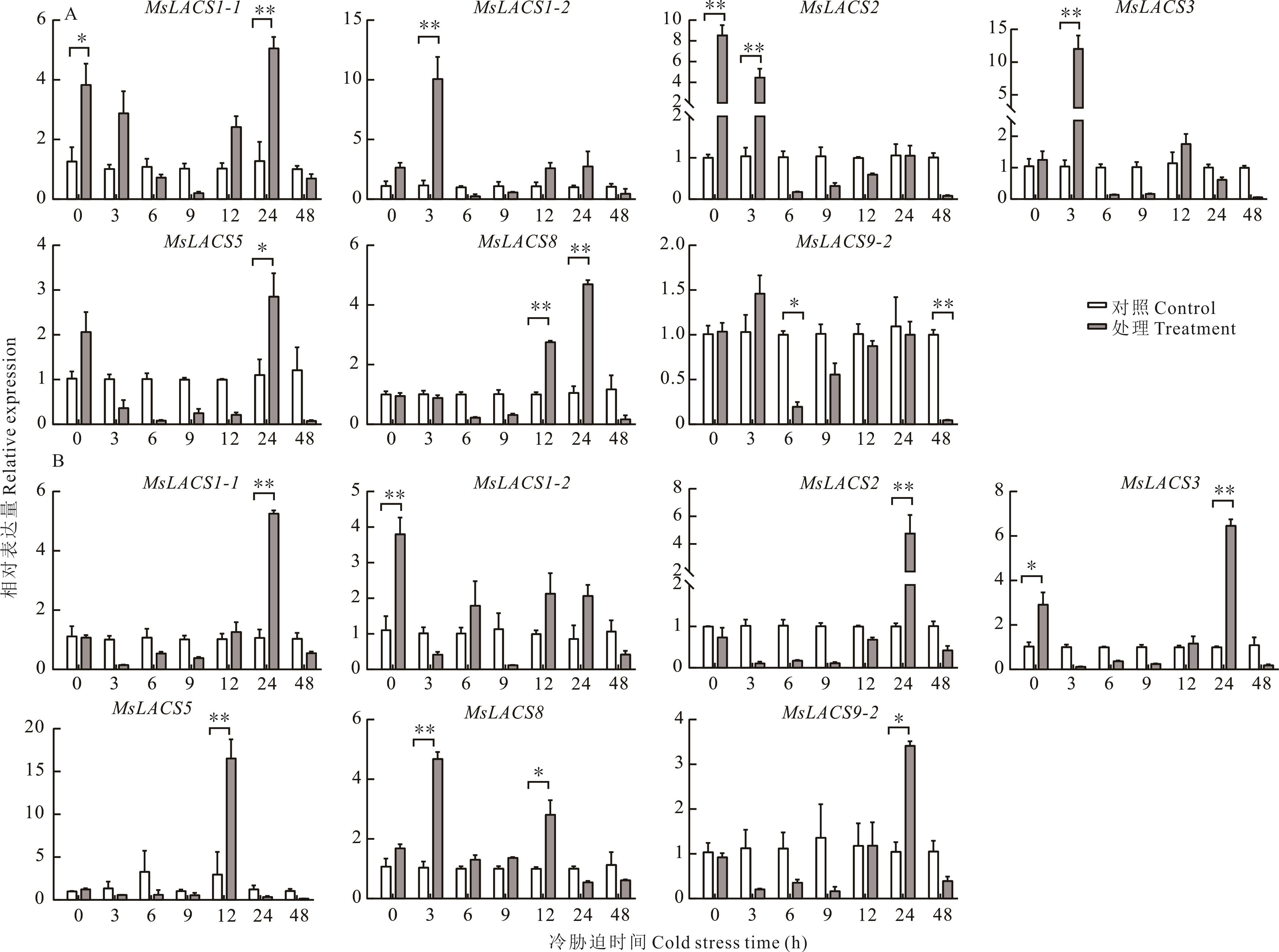
图6 冷胁迫下紫花苜蓿MsLACS基因在叶片和根中的表达A: 叶leaf; B: 根Root. *: P<0.05; **: P<0.01. 下同The same below.
Fig.6 Expression of MsLACS gene in leaves and roots of alfalfa under cold stress
| 1 | Wang Y P, Zeng Y, Luo P. Advances in metabolic engineering of plant fatty acids. Chinese Journal of Oil Crop Science, 1998, 20(4): 88-92. |
| 王幼平, 曾宇, 罗鹏. 植物脂肪酸代谢工程研究进展. 中国油料作物学报, 1998, 20(4): 88-92. | |
| 2 | Li Q G, Tao Z, Yang Y Z, et al. Research progress of long chain acyl-CoA synthetase. China Animal Husbandry and Veterinary Medicine, 2012, 39(6): 137-140. |
| 李庆岗, 陶著, 杨玉增, 等. 长链脂酰CoA合成酶(ACSL)的研究进展. 中国畜牧兽医, 2012, 39(6): 137-140. | |
| 3 | Zhou D, Zhao J Z, Bai Y, et al. Research advance in triacylglycerol synthesis,metabolism, and regulation in plants. Journal of Nanjing Agricultural University, 2012, 35(5): 77-86. |
| 周丹, 赵江哲, 柏杨, 等. 植物油脂合成代谢及调控的研究进展. 南京农业大学学报, 2012, 35(5): 77-86. | |
| 4 | Somerville C, Browse J. Plant lipids: Metabolism, mutants, and membranes. Science, 1991, 252(5002): 80-87. |
| 5 | Hills M J, Beevers H. ATPase in lipid body membranes of castor bean endosperm. Plant Physiology, 1986, 82(3): 671-674. |
| 6 | Mukherjee K D. Plant lipases and their application in lipid bio-transformations. Progress Lipid Research, 1994, 33(1/2): 165-174. |
| 7 | Lv J B, Tan X F, Long H X, et al. Research progress in the study of plant long-chain acyl-coenzyme A(coA). Journal of Plant Physiology, 2017, 53(7): 1185-1191. |
| 吕佳斌, 谭晓风, 龙洪旭, 等. 植物长链脂酰辅酶A合成酶研究进展. 植物生理学报, 2017, 53(7): 1185-1191. | |
| 8 | Shockey J M, Fulda M S, Browse J A. Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiology, 2002, 129(4): 1710-1722. |
| 9 | Tan X L. Characterization of an Arabidopsis long chain fatty acyl-coenzyme A synthetase, which is required for seedling establishment without exogenous sugar. Yangling: Northwest AF University, 2003. |
| 谭小力. 拟南芥长链脂肪酰辅酶A合成酶基因的克隆及功能鉴定. 杨凌: 西北农林科技大学, 2003. | |
| 10 | Lv S Y, Song T, Kosma D K, et al. Arabidopsis CER8 encodes long-chain acyl-CoA synthetase 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal, 2009, 59 (4): 553-564. |
| 11 | Trick H N, Finer J J. Sonication-assisted agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill] embryogenic suspension culture tissue. Plant Cell Reports, 1998, 17(6): 482-488. |
| 12 | Jessen D, Olbrich A, Knüfer J, et al. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis: LACS activity and pollen coat formation. The Plant Journal, 2011, 68(4): 715-726. |
| 13 | Jessen D, Roth C, Wiermer M, et al. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiology, 2015, 167(2): 351-366. |
| 14 | Fulda M, Shockey J, Werber M, et al. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation: Fatty acid activation in peroxisomes of plants. The Plant Journal, 2002, 32(1): 93-103. |
| 15 | Zhao L, Haslam T M, Sonntag A, et al. Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis. Plant and Cell Physiology, 2019, 60(5): 1041-1054. |
| 16 | Dahlqvist A, Stahl U, Lenman M, et al. Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6487-6492. |
| 17 | Visser W F, van Roermund C W, Ijlst L, et al. Metabolite transport across the peroxisomal membrane. Biochemical Journal, 2007, 401(2): 365-375. |
| 18 | Zhang C L, Mao K, Zhou L J, et al. Genome-wide identification and characterization of apple long-chain acyl-CoA synthetases and expression analysis under different stresses. Plant Physiology and Biochemistry, 2018, 132: 320-332. |
| 19 | Black P N, DiRusso C C, Metzger A K, et al. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem, 1993, 267(35): 25513-25520. |
| 20 | Dai H P, Shan C J, Zhao H., et al. The difference in antioxidant capacity of four alfalfa cultivars in response to Zn. Ecotoxicology and Environmental Safety, 2015, 114: 312-317. |
| 21 | Song X, Fang C, Yuan Z Q, et al. Long-term alfalfa (Medicago sativa L.) establishment could alleviate phosphorus limitation induced by nitrogen deposition in the carbonate soil. Journal of Environmental Management, 2022, 324: 116346. |
| 22 | Li Y J, Ma J W, Li Y Q, et al. Effect of nitrogen on the phytoremediation of Cd-PAHs co-contaminated dumpsite soil by alfalfa (Medicago sativa L.) and on the soil bacterial community structure. Environmental Science, 2022, 43(10): 4779-4788. |
| 23 | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2‒ΔΔCt method. Methods, 2001, 25(4): 402-408. |
| 24 | Yang J B, Zhang Z, Zhou Z M, et al. Cloning and function analysis of a HaLACS9 gene in Helianthus annuus L. Acta Agronomica Sinica, 2023, 49(2): 426-437. |
| 杨佳宝, 张展, 周至铭, 等. 向日葵HaLACS9基因的克隆与功能分析. 作物学报, 2023, 49(2): 426-437. | |
| 25 | Deng S Y, Xiao Y, Zhang H T, et al. Bioinformatics and expression analysis of the LACS gene family in Prunus sibirica. Molecular Plant Breeding, 2018, 16(6): 1977-1984. |
| 邓舒雅, 肖羽, 张鹤婷, 等. 山杏LACS基因家族生物信息学及表达分析. 分子植物育种, 2018, 16(6): 1977-1984. | |
| 26 | Jia Q, Liu Y B, Wang F, et al. Identification of LACS family genes in Capsicum annuum L. and their response to abiotic stress. Journal of Yangtze University(Natural Science Edition), 2022, 19(6): 117-126. |
| 贾切, 刘亚博, 王飞, 等. 辣椒中LACS家族基因鉴定及其对非生物胁迫的响应. 长江大学学报(自然科学版), 2022, 19(6): 117-126. | |
| 27 | Wang R H, Wang S B, Liu S T, et al. Identification and expression analysis of LACS family genes in Chinese cabbage (Brassica rapa L.ssp.pekinensis). Shandong Agricultural Sciences, 2022, 54(6): 1-9. |
| 王荣花, 王树彬, 刘栓桃, 等. 大白菜LACS家族基因鉴定与表达分析. 山东农业科学, 2022, 54(6): 1-9. |
| [1] | 马婷, 陈奋奇, 王勇, 哈雪, 李亚君, 马晖玲. NaCl胁迫下鹰嘴紫云英根系基因差异表达及相关通路分析[J]. 草业学报, 2025, 34(4): 104-123. |
| [2] | 陈彩锦, 包明芳, 王文虎, 尚继红, 曾燕霞, 沙晓弟, 朱新忠, 王学敏, 刘文辉. 紫花苜蓿抗旱育种研究现状及展望[J]. 草业学报, 2025, 34(3): 204-223. |
| [3] | 胡鹏飞, 叶雨浓, 王通锐, 王晶, 王星, 伏兵哲, 高雪芹. 紫花苜蓿半同胞家系农艺性状的遗传变异分析[J]. 草业学报, 2025, 34(3): 85-96. |
| [4] | 汪欣瑶, 彭亚萍, 姚立蓉, 汪军成, 司二静, 张宏, 杨轲, 马小乐, 孟亚雄, 王化俊, 李葆春. 盐生草HgS5基因的克隆与抗旱性鉴定[J]. 草业学报, 2025, 34(2): 184-195. |
| [5] | 马超, 孙熙婧, 冯雅岚, 周爽, 琚吉浩, 吴毅, 王添宁, 郭彬彬, 张均. 紫花苜蓿GLK基因家族鉴定及渗透胁迫下的表达分析[J]. 草业学报, 2025, 34(1): 174-190. |
| [6] | 蔡文祺, 李淑霞, 王晓彤, 宋文学, 麻旭霞, 马小梅, 李小红, 代昕瑶. 外源褪黑素与乙烯交互对盐胁迫下紫花苜蓿幼苗生长和生理特性的影响[J]. 草业学报, 2025, 34(1): 80-93. |
| [7] | 崔红丽, 孙明哲, 贾博为, 孙晓丽. 蒺藜苜蓿OSCA基因家族鉴定及低温逆境表达分析[J]. 草业学报, 2024, 33(9): 111-125. |
| [8] | 王晓彤, 李小红, 麻旭霞, 蔡文祺, 冯学丽, 李淑霞. 紫花苜蓿FBA基因家族成员的鉴定与分析[J]. 草业学报, 2024, 33(9): 81-93. |
| [9] | 张盈盈, 胡丹丹, 马春晖, 张前兵. 苜蓿叶片结构和光合特性对菌磷添加的响应[J]. 草业学报, 2024, 33(8): 133-144. |
| [10] | 马圆, 刘欢, 赵桂琴, 王敬龙, 张然, 姚瑞瑞. 燕麦sHSP基因家族的鉴定及其响应高温及老化的表达分析[J]. 草业学报, 2024, 33(8): 145-158. |
| [11] | 王峥, 常伟, 李俊诚, 苏连泰, 高鲤, 周鹏, 安渊. 紫花苜蓿还田对饲料玉米产量和氮素吸收转运的影响[J]. 草业学报, 2024, 33(8): 63-73. |
| [12] | 吴毅, 冯雅岚, 王添宁, 琚吉浩, 肖慧淑, 马超, 张均. 小麦及其祖先物种Hsp70基因家族鉴定与表达分析[J]. 草业学报, 2024, 33(7): 53-67. |
| [13] | 张震欢, 姚立蓉, 汪军成, 司二静, 张宏, 杨轲, 马小乐, 孟亚雄, 王化俊, 李葆春. 盐生草AKR基因家族成员的鉴定及根系盐胁迫响应基因HgAKR42639的耐盐分析[J]. 草业学报, 2024, 33(7): 68-83. |
| [14] | 宋淑珍, 朱才业, 刘立山, 宫旭胤, 雒瑞瑞. 断尾对兰州大尾羊脂肪细胞结构和脂肪代谢相关基因表达的影响[J]. 草业学报, 2024, 33(7): 94-104. |
| [15] | 高金柱, 赵东豪, 高乐, 苏喜浩, 何学青. 硝酸铈与脱落酸处理对紫花苜蓿种子萌发和幼苗生理特性的影响[J]. 草业学报, 2024, 33(6): 175-186. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||