

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2025, Vol. 34 ›› Issue (1): 174-190.DOI: 10.11686/cyxb2024133
• 研究论文 • 上一篇
马超1( ), 孙熙婧1(
), 孙熙婧1( ), 冯雅岚2, 周爽1, 琚吉浩1, 吴毅1, 王添宁1, 郭彬彬1, 张均1(
), 冯雅岚2, 周爽1, 琚吉浩1, 吴毅1, 王添宁1, 郭彬彬1, 张均1( )
)
收稿日期:2024-04-18
修回日期:2024-06-05
出版日期:2025-01-20
发布日期:2024-11-04
通讯作者:
张均
作者简介:E-mail: zhangjun0105@126.com基金资助:
Chao MA1( ), Xi-jing SUN1(
), Xi-jing SUN1( ), Ya-lan FENG2, Shuang ZHOU1, Ji-hao JU1, Yi WU1, Tian-ning WANG1, Bin-bin GUO1, Jun ZHANG1(
), Ya-lan FENG2, Shuang ZHOU1, Ji-hao JU1, Yi WU1, Tian-ning WANG1, Bin-bin GUO1, Jun ZHANG1( )
)
Received:2024-04-18
Revised:2024-06-05
Online:2025-01-20
Published:2024-11-04
Contact:
Jun ZHANG
About author:First author contact:These authors contributed equally to this work.
摘要:
GLK (Golden2-like或G2-like)属于GARP超家族,是植物生长发育过程中的重要转录因子,在调节植物叶绿体发育、叶绿素生物合成和非生物胁迫响应等方面发挥着重要作用。目前,GLK基因家族成员已在多个物种中被系统鉴定,但在四倍体紫花苜蓿全基因组水平上仍然知之甚少。利用生物信息学的方法,在“新疆大叶”紫花苜蓿基因组中鉴定到100个GLK基因(MsGLKs),并对其理化性质、染色体定位、系统进化关系、启动子顺式作用元件以及渗透胁迫和外源脱落酸(ABA)处理下的表达模式进行了分析。结果显示,100个MsGLK基因在32条染色体上不均匀分布,蛋白序列长度为201~860个氨基酸。根据系统发育分析结果,将MsGLK家族成员分为13个组。共线性分析表明,在紫花苜蓿基因组中共发现193个MsGLK基因重复事件,基因非同义替代数/同义替代数(Ka/Ks)分析显示,大部分重复基因对经历了纯化选择。MsGLK基因启动子的顺式作用元件广泛参与了植物生长发育、激素响应和胁迫反应。基因表达数据显示,12个基因的表达具有组织特异性,25个基因在所有组织中表达。RT-qPCR检测发现,MsGLK基因在干旱胁迫、盐胁迫和外源ABA处理下均有一定程度的响应。研究结果将为进一步探索 MsGLK基因的功能和紫花苜蓿抗逆性遗传改良提供参考。
马超, 孙熙婧, 冯雅岚, 周爽, 琚吉浩, 吴毅, 王添宁, 郭彬彬, 张均. 紫花苜蓿GLK基因家族鉴定及渗透胁迫下的表达分析[J]. 草业学报, 2025, 34(1): 174-190.
Chao MA, Xi-jing SUN, Ya-lan FENG, Shuang ZHOU, Ji-hao JU, Yi WU, Tian-ning WANG, Bin-bin GUO, Jun ZHANG. Genome-wide identification of the GLK gene family in alfalfa and their transcript profiles under osmotic stress[J]. Acta Prataculturae Sinica, 2025, 34(1): 174-190.
| 基因Gene | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsGLK10 | CAGATGAAGTCGGATGAGCCT | GGTGGTGCGGTAAGTTGGTC |
| MsGLK17 | TACCACAGCAAGGGAAGCAG | GAATGGAACGGCGATGAACT |
| MsGLK29 | GCAAGTGGAGGCAGAGAAGC | GGGTGGGATGGTTTGAGGA |
| MsGLK33 | AAACCTGCCTCTTTACCGATTC | GTTGCTCTTTCTTGCCCTCC |
| MsGLK36 | GCAAGTGGAGGCAGAGAAGC | GGGTGGGATGGTTTGAGGA |
| MsGLK62 | CATCACTCATCCCTTTCACCTC | CCTGCCAATCATTTCGCTT |
| MsGLK65 | AACACTTCCGCTGTCCCAT | TCTCATTGCCGTCGTCATTT |
| MsGLK75 | CTTCATCACTCATCCCTTTCACC | GCAGGCACCGCTCTCTACTT |
| MsGLK83 | ATATCCAGGGTCATAGTATTTTCCA | CGTTGTTTTCAGTTCTTCCTCTTG |
| MsActin | ACTGGAATGGTGAAGGCTGG | TGACAATACCGTGCTCAATGG |
表1 RT-qPCR引物信息
Table 1 Primers used for RT-qPCR
| 基因Gene | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| MsGLK10 | CAGATGAAGTCGGATGAGCCT | GGTGGTGCGGTAAGTTGGTC |
| MsGLK17 | TACCACAGCAAGGGAAGCAG | GAATGGAACGGCGATGAACT |
| MsGLK29 | GCAAGTGGAGGCAGAGAAGC | GGGTGGGATGGTTTGAGGA |
| MsGLK33 | AAACCTGCCTCTTTACCGATTC | GTTGCTCTTTCTTGCCCTCC |
| MsGLK36 | GCAAGTGGAGGCAGAGAAGC | GGGTGGGATGGTTTGAGGA |
| MsGLK62 | CATCACTCATCCCTTTCACCTC | CCTGCCAATCATTTCGCTT |
| MsGLK65 | AACACTTCCGCTGTCCCAT | TCTCATTGCCGTCGTCATTT |
| MsGLK75 | CTTCATCACTCATCCCTTTCACC | GCAGGCACCGCTCTCTACTT |
| MsGLK83 | ATATCCAGGGTCATAGTATTTTCCA | CGTTGTTTTCAGTTCTTCCTCTTG |
| MsActin | ACTGGAATGGTGAAGGCTGG | TGACAATACCGTGCTCAATGG |
基因名 Gene name | 基因登录号 Gene ID | PL (aa) | Mw (kDa) | pI | Sl | 基因名 Gene name | 基因登录号 Gene ID | PL (aa) | Mw (kDa) | pI | Sl |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MsGLK1 | MS.gene005086.t1 | 388 | 43.19 | 4.73 | Nuc | MsGLK51 | MS.gene76602.t1 | 352 | 39.86 | 5.75 | Nuc |
| MsGLK2 | MS.gene005866.t1 | 378 | 42.16 | 7.72 | Nuc | MsGLK52 | MS.gene55246.t1 | 352 | 40.14 | 5.54 | Nuc |
| MsGLK3 | MS.gene041038.t1 | 281 | 32.18 | 8.66 | Nuc | MsGLK53 | MS.gene028039.t1 | 277 | 31.24 | 6.03 | Nuc |
| MsGLK4 | MS.gene37289.t1 | 231 | 26.06 | 8.63 | Nuc | MsGLK54 | MS.gene045645.t1 | 353 | 40.08 | 5.75 | Nuc |
| MsGLK5 | MS.gene60590.t1 | 379 | 42.20 | 7.18 | Nuc | MsGLK55 | MS.gene070117.t1 | 277 | 31.32 | 5.82 | Nuc |
| MsGLK6 | MS.gene43963.t1 | 388 | 43.13 | 4.76 | Nuc | MsGLK56 | MS.gene032930.t1 | 352 | 39.83 | 5.75 | Nuc |
| MsGLK7 | MS.gene001686.t1 | 378 | 42.16 | 7.73 | Nuc | MsGLK57 | MS.gene87987.t1 | 393 | 44.55 | 6.20 | Nuc |
| MsGLK8 | MS.gene070698.t1 | 437 | 48.03 | 6.98 | Nuc | MsGLK58 | MS.gene85671.t1 | 308 | 34.34 | 9.37 | Nuc |
| MsGLK9 | MS.gene026358.t1 | 201 | 23.51 | 8.81 | Nuc | MsGLK59 | MS.gene68923.t1 | 393 | 44.48 | 6.16 | Nuc |
| MsGLK10 | MS.gene036427.t1 | 388 | 43.13 | 4.76 | Nuc | MsGLK60 | MS.gene042587.t1 | 393 | 44.45 | 6.07 | Nuc |
| MsGLK11 | MS.gene21397.t1 | 297 | 33.01 | 6.43 | Nuc | MsGLK61 | MS.gene66964.t1 | 393 | 44.48 | 6.16 | Nuc |
| MsGLK12 | MS.gene60862.t1 | 378 | 42.15 | 7.72 | Nuc | MsGLK62 | MS.gene018653.t1 | 489 | 53.80 | 5.42 | Nuc |
| MsGLK13 | MS.gene055776.t1 | 423 | 46.76 | 5.86 | Nuc | MsGLK63 | MS.gene017974.t1 | 359 | 40.43 | 6.92 | Nuc |
| MsGLK14 | MS.gene35408.t1 | 373 | 41.81 | 7.74 | Nuc | MsGLK64 | MS.gene017688.t1 | 301 | 32.94 | 6.56 | Nuc |
| MsGLK15 | MS.gene35289.t1 | 257 | 28.82 | 8.91 | Nuc | MsGLK65 | MS.gene013905.t1 | 281 | 30.65 | 5.79 | Nuc |
| MsGLK16 | MS.gene62480.t1 | 365 | 40.90 | 8.66 | Cyt | MsGLK66 | MS.gene99484.t1 | 350 | 39.92 | 5.48 | Nuc |
| MsGLK17 | MS.gene003164.t1 | 860 | 94.37 | 8.47 | Chl | MsGLK67 | MS.gene84188.t1 | 238 | 26.82 | 9.64 | Nuc |
| MsGLK18 | MS.gene003165.t1 | 330 | 36.16 | 6.19 | Nuc | MsGLK68 | MS.gene035191.t1 | 489 | 53.78 | 5.42 | Nuc |
| MsGLK19 | MS.gene043457.t1 | 423 | 46.73 | 5.86 | Nuc | MsGLK69 | MS.gene52648.t1 | 351 | 39.53 | 6.70 | Nuc |
| MsGLK20 | MS.gene84326.t1 | 373 | 41.81 | 7.74 | Nuc | MsGLK70 | MS.gene013966.t1 | 301 | 32.84 | 6.56 | Nuc |
| MsGLK21 | MS.gene073152.t1 | 313 | 35.17 | 6.98 | Nuc | MsGLK71 | MS.gene068055.t1 | 283 | 30.79 | 5.94 | Nuc |
| MsGLK22 | MS.gene066206.t1 | 375 | 41.71 | 8.37 | Nuc | MsGLK72 | MS.gene069022.t1 | 350 | 39.82 | 5.48 | Nuc |
| MsGLK23 | MS.gene01976.t1 | 331 | 36.21 | 6.23 | Nuc | MsGLK73 | MS.gene77355.t1 | 256 | 28.91 | 9.60 | Nuc |
| MsGLK24 | MS.gene01975.t1 | 382 | 41.62 | 8.62 | Chl | MsGLK74 | MS.gene024551.t1 | 439 | 48.79 | 5.11 | Nuc |
| MsGLK25 | MS.gene028866.t1 | 423 | 46.73 | 5.86 | Nuc | MsGLK75 | MS.gene23439.t1 | 489 | 53.78 | 5.42 | Nuc |
| MsGLK26 | MS.gene034456.t1 | 373 | 41.83 | 7.25 | Nuc | MsGLK76 | MS.gene04612.t1 | 359 | 40.43 | 6.92 | Nuc |
| MsGLK27 | MS.gene040544.t1 | 581 | 63.91 | 5.80 | Nuc | MsGLK77 | MS.gene58831.t1 | 298 | 32.76 | 6.56 | Nuc |
| MsGLK28 | MS.gene045711.t1 | 375 | 41.72 | 8.37 | Nuc | MsGLK78 | MS.gene77413.t1 | 350 | 39.94 | 5.56 | Nuc |
| MsGLK29 | MS.gene048293.t1 | 314 | 34.41 | 6.60 | Nuc | MsGLK79 | MS.gene94669.t1 | 256 | 28.93 | 9.78 | Nuc |
| MsGLK30 | MS.gene048294.t1 | 330 | 36.16 | 6.19 | Nuc | MsGLK80 | MS.gene050550.t1 | 439 | 48.79 | 5.11 | Nuc |
| MsGLK31 | MS.gene31432.t1 | 423 | 46.69 | 5.86 | Nuc | MsGLK81 | MS.gene96951.t1 | 489 | 53.73 | 5.37 | Nuc |
| MsGLK32 | MS.gene43040.t1 | 373 | 41.79 | 7.25 | Nuc | MsGLK82 | MS.gene41865.t1 | 283 | 30.79 | 5.94 | Nuc |
| MsGLK33 | MS.gene94587.t1 | 315 | 35.25 | 7.30 | Nuc | MsGLK83 | MS.gene95656.t1 | 350 | 39.85 | 5.48 | Nuc |
| MsGLK34 | MS.gene038958.t1 | 375 | 41.74 | 8.37 | Nuc | MsGLK84 | MS.gene95601.t1 | 238 | 26.83 | 9.64 | Nuc |
| MsGLK35 | MS.gene02861.t1 | 330 | 36.17 | 6.24 | Nuc | MsGLK85 | MS.gene036866.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK36 | MS.gene02867.t1 | 314 | 34.42 | 6.60 | Nuc | MsGLK86 | MS.gene61025.t1 | 352 | 39.45 | 6.50 | Nuc |
| MsGLK37 | MS.gene002965.t1 | 238 | 26.64 | 8.87 | Nuc | MsGLK87 | MS.gene068940.t1 | 345 | 38.77 | 6.64 | Nuc |
| MsGLK38 | MS.gene008318.t1 | 461 | 50.91 | 7.22 | Nuc | MsGLK88 | MS.gene060554.t1 | 434 | 47.88 | 6.66 | Nuc |
| MsGLK39 | MS.gene019228.t1 | 383 | 42.69 | 7.20 | Chl | MsGLK89 | MS.gene42755.t1 | 345 | 38.77 | 6.64 | Nuc |
| MsGLK40 | MS.gene061558.t1 | 426 | 47.13 | 7.99 | Nuc | MsGLK90 | MS.gene36713.t1 | 221 | 25.38 | 9.67 | Chl |
| MsGLK41 | MS.gene057902.t1 | 394 | 43.82 | 9.10 | Nuc | MsGLK91 | MS.gene87427.t1 | 650 | 71.83 | 5.55 | Nuc |
| MsGLK42 | MS.gene049714.t1 | 424 | 46.87 | 7.99 | Nuc | MsGLK92 | MS.gene36099.t1 | 346 | 38.76 | 6.64 | Nuc |
| MsGLK43 | MS.gene05012.t1 | 425 | 47.00 | 7.99 | Nuc | MsGLK93 | MS.gene88392.t1 | 302 | 33.85 | 8.12 | Cyt |
| MsGLK44 | MS.gene26549.t1 | 420 | 47.03 | 8.17 | Nuc | MsGLK94 | MS.gene056957.t1 | 363 | 40.53 | 9.07 | Nuc |
| MsGLK45 | MS.gene006790.t1 | 340 | 37.99 | 5.97 | Nuc | MsGLK95 | MS.gene056956.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK46 | MS.gene031499.t1 | 340 | 38.06 | 5.97 | Nuc | MsGLK96 | MS.gene038713.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK47 | MS.gene96791.t1 | 340 | 37.99 | 5.97 | Nuc | MsGLK97 | MS.gene066880.t1 | 346 | 38.90 | 6.68 | Nuc |
| MsGLK48 | MS.gene56537.t1 | 339 | 37.89 | 5.97 | Nuc | MsGLK98 | MS.gene00817.t1 | 238 | 26.61 | 8.87 | Nuc |
| MsGLK49 | MS.gene54226.t1 | 387 | 43.44 | 7.27 | Nuc | MsGLK99 | MS.gene02381.t1 | 238 | 26.66 | 8.87 | Nuc |
| MsGLK50 | MS.gene72865.t1 | 277 | 31.26 | 5.95 | Nuc | MsGLK100 | MS.gene072393.t1 | 351 | 39.62 | 6.73 | Nuc |
表2 紫花苜蓿GLK家族的基本信息分析
Table 2 Basic information analysis of GLK gene family of alfalfa
基因名 Gene name | 基因登录号 Gene ID | PL (aa) | Mw (kDa) | pI | Sl | 基因名 Gene name | 基因登录号 Gene ID | PL (aa) | Mw (kDa) | pI | Sl |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MsGLK1 | MS.gene005086.t1 | 388 | 43.19 | 4.73 | Nuc | MsGLK51 | MS.gene76602.t1 | 352 | 39.86 | 5.75 | Nuc |
| MsGLK2 | MS.gene005866.t1 | 378 | 42.16 | 7.72 | Nuc | MsGLK52 | MS.gene55246.t1 | 352 | 40.14 | 5.54 | Nuc |
| MsGLK3 | MS.gene041038.t1 | 281 | 32.18 | 8.66 | Nuc | MsGLK53 | MS.gene028039.t1 | 277 | 31.24 | 6.03 | Nuc |
| MsGLK4 | MS.gene37289.t1 | 231 | 26.06 | 8.63 | Nuc | MsGLK54 | MS.gene045645.t1 | 353 | 40.08 | 5.75 | Nuc |
| MsGLK5 | MS.gene60590.t1 | 379 | 42.20 | 7.18 | Nuc | MsGLK55 | MS.gene070117.t1 | 277 | 31.32 | 5.82 | Nuc |
| MsGLK6 | MS.gene43963.t1 | 388 | 43.13 | 4.76 | Nuc | MsGLK56 | MS.gene032930.t1 | 352 | 39.83 | 5.75 | Nuc |
| MsGLK7 | MS.gene001686.t1 | 378 | 42.16 | 7.73 | Nuc | MsGLK57 | MS.gene87987.t1 | 393 | 44.55 | 6.20 | Nuc |
| MsGLK8 | MS.gene070698.t1 | 437 | 48.03 | 6.98 | Nuc | MsGLK58 | MS.gene85671.t1 | 308 | 34.34 | 9.37 | Nuc |
| MsGLK9 | MS.gene026358.t1 | 201 | 23.51 | 8.81 | Nuc | MsGLK59 | MS.gene68923.t1 | 393 | 44.48 | 6.16 | Nuc |
| MsGLK10 | MS.gene036427.t1 | 388 | 43.13 | 4.76 | Nuc | MsGLK60 | MS.gene042587.t1 | 393 | 44.45 | 6.07 | Nuc |
| MsGLK11 | MS.gene21397.t1 | 297 | 33.01 | 6.43 | Nuc | MsGLK61 | MS.gene66964.t1 | 393 | 44.48 | 6.16 | Nuc |
| MsGLK12 | MS.gene60862.t1 | 378 | 42.15 | 7.72 | Nuc | MsGLK62 | MS.gene018653.t1 | 489 | 53.80 | 5.42 | Nuc |
| MsGLK13 | MS.gene055776.t1 | 423 | 46.76 | 5.86 | Nuc | MsGLK63 | MS.gene017974.t1 | 359 | 40.43 | 6.92 | Nuc |
| MsGLK14 | MS.gene35408.t1 | 373 | 41.81 | 7.74 | Nuc | MsGLK64 | MS.gene017688.t1 | 301 | 32.94 | 6.56 | Nuc |
| MsGLK15 | MS.gene35289.t1 | 257 | 28.82 | 8.91 | Nuc | MsGLK65 | MS.gene013905.t1 | 281 | 30.65 | 5.79 | Nuc |
| MsGLK16 | MS.gene62480.t1 | 365 | 40.90 | 8.66 | Cyt | MsGLK66 | MS.gene99484.t1 | 350 | 39.92 | 5.48 | Nuc |
| MsGLK17 | MS.gene003164.t1 | 860 | 94.37 | 8.47 | Chl | MsGLK67 | MS.gene84188.t1 | 238 | 26.82 | 9.64 | Nuc |
| MsGLK18 | MS.gene003165.t1 | 330 | 36.16 | 6.19 | Nuc | MsGLK68 | MS.gene035191.t1 | 489 | 53.78 | 5.42 | Nuc |
| MsGLK19 | MS.gene043457.t1 | 423 | 46.73 | 5.86 | Nuc | MsGLK69 | MS.gene52648.t1 | 351 | 39.53 | 6.70 | Nuc |
| MsGLK20 | MS.gene84326.t1 | 373 | 41.81 | 7.74 | Nuc | MsGLK70 | MS.gene013966.t1 | 301 | 32.84 | 6.56 | Nuc |
| MsGLK21 | MS.gene073152.t1 | 313 | 35.17 | 6.98 | Nuc | MsGLK71 | MS.gene068055.t1 | 283 | 30.79 | 5.94 | Nuc |
| MsGLK22 | MS.gene066206.t1 | 375 | 41.71 | 8.37 | Nuc | MsGLK72 | MS.gene069022.t1 | 350 | 39.82 | 5.48 | Nuc |
| MsGLK23 | MS.gene01976.t1 | 331 | 36.21 | 6.23 | Nuc | MsGLK73 | MS.gene77355.t1 | 256 | 28.91 | 9.60 | Nuc |
| MsGLK24 | MS.gene01975.t1 | 382 | 41.62 | 8.62 | Chl | MsGLK74 | MS.gene024551.t1 | 439 | 48.79 | 5.11 | Nuc |
| MsGLK25 | MS.gene028866.t1 | 423 | 46.73 | 5.86 | Nuc | MsGLK75 | MS.gene23439.t1 | 489 | 53.78 | 5.42 | Nuc |
| MsGLK26 | MS.gene034456.t1 | 373 | 41.83 | 7.25 | Nuc | MsGLK76 | MS.gene04612.t1 | 359 | 40.43 | 6.92 | Nuc |
| MsGLK27 | MS.gene040544.t1 | 581 | 63.91 | 5.80 | Nuc | MsGLK77 | MS.gene58831.t1 | 298 | 32.76 | 6.56 | Nuc |
| MsGLK28 | MS.gene045711.t1 | 375 | 41.72 | 8.37 | Nuc | MsGLK78 | MS.gene77413.t1 | 350 | 39.94 | 5.56 | Nuc |
| MsGLK29 | MS.gene048293.t1 | 314 | 34.41 | 6.60 | Nuc | MsGLK79 | MS.gene94669.t1 | 256 | 28.93 | 9.78 | Nuc |
| MsGLK30 | MS.gene048294.t1 | 330 | 36.16 | 6.19 | Nuc | MsGLK80 | MS.gene050550.t1 | 439 | 48.79 | 5.11 | Nuc |
| MsGLK31 | MS.gene31432.t1 | 423 | 46.69 | 5.86 | Nuc | MsGLK81 | MS.gene96951.t1 | 489 | 53.73 | 5.37 | Nuc |
| MsGLK32 | MS.gene43040.t1 | 373 | 41.79 | 7.25 | Nuc | MsGLK82 | MS.gene41865.t1 | 283 | 30.79 | 5.94 | Nuc |
| MsGLK33 | MS.gene94587.t1 | 315 | 35.25 | 7.30 | Nuc | MsGLK83 | MS.gene95656.t1 | 350 | 39.85 | 5.48 | Nuc |
| MsGLK34 | MS.gene038958.t1 | 375 | 41.74 | 8.37 | Nuc | MsGLK84 | MS.gene95601.t1 | 238 | 26.83 | 9.64 | Nuc |
| MsGLK35 | MS.gene02861.t1 | 330 | 36.17 | 6.24 | Nuc | MsGLK85 | MS.gene036866.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK36 | MS.gene02867.t1 | 314 | 34.42 | 6.60 | Nuc | MsGLK86 | MS.gene61025.t1 | 352 | 39.45 | 6.50 | Nuc |
| MsGLK37 | MS.gene002965.t1 | 238 | 26.64 | 8.87 | Nuc | MsGLK87 | MS.gene068940.t1 | 345 | 38.77 | 6.64 | Nuc |
| MsGLK38 | MS.gene008318.t1 | 461 | 50.91 | 7.22 | Nuc | MsGLK88 | MS.gene060554.t1 | 434 | 47.88 | 6.66 | Nuc |
| MsGLK39 | MS.gene019228.t1 | 383 | 42.69 | 7.20 | Chl | MsGLK89 | MS.gene42755.t1 | 345 | 38.77 | 6.64 | Nuc |
| MsGLK40 | MS.gene061558.t1 | 426 | 47.13 | 7.99 | Nuc | MsGLK90 | MS.gene36713.t1 | 221 | 25.38 | 9.67 | Chl |
| MsGLK41 | MS.gene057902.t1 | 394 | 43.82 | 9.10 | Nuc | MsGLK91 | MS.gene87427.t1 | 650 | 71.83 | 5.55 | Nuc |
| MsGLK42 | MS.gene049714.t1 | 424 | 46.87 | 7.99 | Nuc | MsGLK92 | MS.gene36099.t1 | 346 | 38.76 | 6.64 | Nuc |
| MsGLK43 | MS.gene05012.t1 | 425 | 47.00 | 7.99 | Nuc | MsGLK93 | MS.gene88392.t1 | 302 | 33.85 | 8.12 | Cyt |
| MsGLK44 | MS.gene26549.t1 | 420 | 47.03 | 8.17 | Nuc | MsGLK94 | MS.gene056957.t1 | 363 | 40.53 | 9.07 | Nuc |
| MsGLK45 | MS.gene006790.t1 | 340 | 37.99 | 5.97 | Nuc | MsGLK95 | MS.gene056956.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK46 | MS.gene031499.t1 | 340 | 38.06 | 5.97 | Nuc | MsGLK96 | MS.gene038713.t1 | 434 | 47.87 | 6.66 | Nuc |
| MsGLK47 | MS.gene96791.t1 | 340 | 37.99 | 5.97 | Nuc | MsGLK97 | MS.gene066880.t1 | 346 | 38.90 | 6.68 | Nuc |
| MsGLK48 | MS.gene56537.t1 | 339 | 37.89 | 5.97 | Nuc | MsGLK98 | MS.gene00817.t1 | 238 | 26.61 | 8.87 | Nuc |
| MsGLK49 | MS.gene54226.t1 | 387 | 43.44 | 7.27 | Nuc | MsGLK99 | MS.gene02381.t1 | 238 | 26.66 | 8.87 | Nuc |
| MsGLK50 | MS.gene72865.t1 | 277 | 31.26 | 5.95 | Nuc | MsGLK100 | MS.gene072393.t1 | 351 | 39.62 | 6.73 | Nuc |

图6 紫花苜蓿GLK基因在不同组织中的表达分析A~F: 在1~6种组织中MsGLK基因的表达The expression of MsGLK gene in 1-6 tissues. 热图上方的色阶表示相对表达水平,从白色到红色的颜色渐变表示表达水平的增加The color scale above the heatmap indicates the relative expression levels, and the color gradient from white to red indicates an increase in expression levels. G: 在0~6种组织中表达的MsGLK基因数量Number of MsGLK genes expressed in 0-6 tissues. 下同The same below.
Fig.6 Analysis of GLK gene expression in different tissues of alfalfa
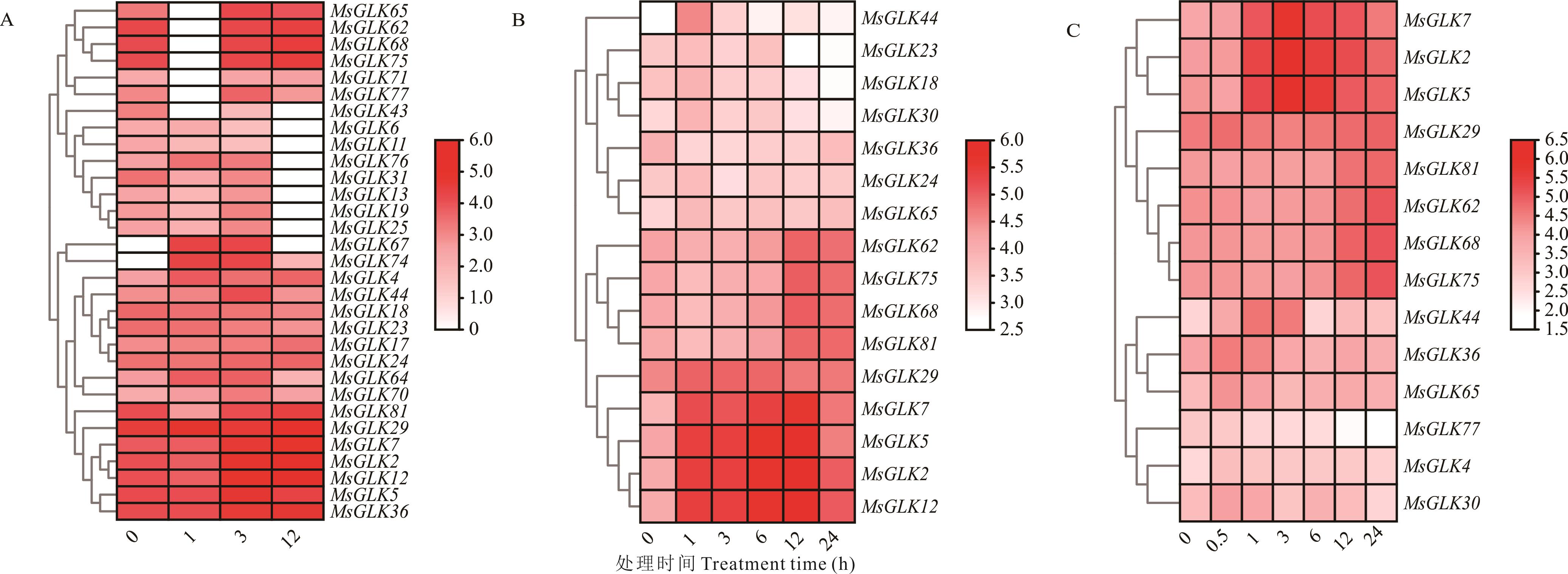
图7 紫花苜蓿GLK基因在渗透胁迫下的表达分析A: 10 μmol·L-1脱落酸ABA; B: 400 mmol·L-1甘露醇Mannitol; C: 250 mmol·L-1盐处理NaCl treatment.
Fig.7 Expression analysis of GLK genes in alfalfa under osmotic stress

图8 MsGLKs基因家族成员在外源ABA和渗透胁迫下的表达模式分析*: 同一时间各胁迫处理与对照差异显著(P<0.05) At the same time, there were significant differences among stress treatment and CK (P<0.05).
Fig.8 Expression patterns of MsGLKs gene family members under exogenous ABA and osmotic stress
| 1 | Zaikina E A, Rumyantsev S D, Sarvarova E R, et al. Transcription factor genes involved in plant response to abiotic stress factors. Ecological Genetics, 2019, 17(3): 47-58. |
| 2 | Joshi R, Wani S H, Singh B, et al. Transcription factors and plants response to drought stress: Current understanding and future directions. Frontiers in Plant Science, 2016, 7: 1029. |
| 3 | Riechmann J L, Heard J, Martin G, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 2000, 290(5499): 2105-2110. |
| 4 | Yasumura Y, Moylan E C, Langdale J A. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. The Plant Cell, 2005, 17(7): 1894-1907. |
| 5 | Rossini L, Cribb L, Martin D J, et al. The maize Golden2 gene defines a novel class of transcriptional regulators in plants. The Plant Cell, 2001, 13(5): 1231-1244. |
| 6 | Tachibana R, Abe S, Marugami M, et al. BPG4 regulates chloroplast development and homeostasis by suppressing GLK transcription factors and involving light and brassinosteroid signaling. Nature Communications, 2024, 15(1): 370. |
| 7 | Waters M T, Wang P, Korkaric M, et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. The Plant Cell, 2009, 21(4): 1109-1128. |
| 8 | Cackett L, Luginbuehl L H, Schreier T B, et al. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytologist, 2022, 233(5): 2000-2016. |
| 9 | Wang P, Fouracre J, Kelly S, et al. Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta, 2013, 237(2):481-495. |
| 10 | Liu X Y, Chen X, Cao J J, et al. GOLDEN 2-LIKE transcription factors regulate chlorophyll biosynthesis and flavonoid accumulation in response to UV-B in tea plants. Horticultural Plant Journal, 2023, 9(5): 1055-1066. |
| 11 | Tu X Y, Ren S B, Shen W, et al. Limited conservation in cross-species comparison of GLK transcription factor binding suggested wide-spread cistrome divergence. Nature Communications, 2022, 13(1): 7632. |
| 12 | Powell A L, Nguyen C V, Hill T, et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science, 2012, 336(6089): 1711-1715. |
| 13 | Li H, Li Y X, Deng H, et al. Tomato UV-B receptor SlUVR8 mediates plant acclimation to UV-B radiation and enhances fruit chloroplast development via regulating SlGLK2. Scientific Reports, 2018, 8(1): 6097. |
| 14 | Zubo Y O, Blakley I C, Franco-Zorrilla J M, et al. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiology, 2018, 178(1): 130-147. |
| 15 | Kim S, Choi H, Yi T, et al. Pleiotropic properties of GOLDEN2-LIKE transcription factors for crop improvement. Applied Biological Chemistry, 2023, 66(1): 1-14. |
| 16 | Singh J, Savitch L, Subramaniam G, et al. The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochemical and Biophysical Research Communications, 2007, 359(2): 234-238. |
| 17 | Han X Y, Li P X, Zou L J, et al. GOLDEN 2-LIKE transcription factors coordinate the tolerance to cucumber mosaic virus in Arabidopsis. Biochemical and Biophysical Research Communications, 2016, 477(4): 626-632. |
| 18 | Ali N, Chen H, Zhang C, et al. Ectopic expression of AhGLK1b (GOLDEN2-like transcription factor) in Arabidopsis confers dual resistance to fungal and bacterial pathogens. Genes, 2020, 11(3): 343. |
| 19 | Townsend P D, Dixon C H, Slootweg E J, et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. Journal of Biological Chemistry, 2018, 293(9): 3218-3233. |
| 20 | Liu J N, Mehari T G, Xu Y C, et al. GhGLK1 a key candidate gene from GARP family enhances cold and drought stress tolerance in cotton. Frontiers in Plant Science, 2021, 12: 759312. |
| 21 | Liu X, Li L M, Li M J, et al. AhGLK1 affects chlorophyll biosynthesis and photosynthesis in peanut leaves during recovery from drought. Scientific Reports, 2018, 8(1): 2250. |
| 22 | Liu F, Xu Y X, Han G M, et al. Molecular evolution and genetic variation of G2-like transcription factor genes in maize. PLoS One, 2016, 11(8): e0161763. |
| 23 | Li T T. Expression analysis of PPO, GLK1 and NADH in lotus and identification of its interaction domain. Yangling: Northwest A & F University, 2021. |
| 李婷婷. 莲藕PPO与GLK1、NADH的表达分析及互作结构域确定. 杨凌: 西北农林科技大学, 2021. | |
| 24 | Ahmad R, Liu Y T, Wang T J, et al. GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiology, 2019, 179(4): 1844-1860. |
| 25 | Chen L, He F, Long R C, et al. A global alfalfa diversity panel reveals genomic selection signatures in Chinese varieties and genomic associations with root development. Journal of Integrative Plant Biology, 2021, 63(11): 1937-1951. |
| 26 | Huang X F, Shi P L, Yu C Q, et al. A review of abiotic stress resistance of forages. Acta Agrestia Sinica, 2023, 31(5): 1293-1301. |
| 黄小芳, 石培礼, 余成群, 等. 非生物胁迫下牧草抗逆性研究进展. 草地学报, 2023, 31(5): 1293-1301. | |
| 27 | Jia Y K, Gao H H, Feng J C, et al. Genome-wide identification and expression analysis of G2-like transcription factors family gene in wheat. Acta Agronomica Sinica, 2023, 49(5): 1410-1425. |
| 贾玉库, 高宏欢, 冯健超, 等. 小麦G2-like转录因子家族基因鉴定与表达模式分析. 作物学报, 2023, 49(5): 1410-1425. | |
| 28 | Wang Z Y, Zhao S, Liu J F, et al. Genome-wide identification of tomato Golden 2-like transcription factors and abiotic stress related members screening. BMC Plant Biology, 2022, 22(1): 82. |
| 29 | Chen H F, Qin L, Tian J G, et al. Identification and evolutionary analysis of the GOLDEN 2-LIKE gene family in foxtail millet. Tropical Plant Biology, 2022, 15: 301-318. |
| 30 | Alam I, Manghwar H, Zhang H Y, et al. Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Frontiers in Plant Science, 2022, 13: 1052659. |
| 31 | Qin M Y, Zhang B H, Gu G, et al. Genome-wide analysis of the G2-like transcription factor genes and their expression in different senescence stages of tobacco (Nicotiana tabacum L.). Frontiers in Genetics, 2021, 31(12): 626352. |
| 32 | Zhao Z L, Shuang J R, Li Z G, et al. Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ, 2021, 9: e12484. |
| 33 | Chen C J, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 34 | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| 35 | O’Rourke J A, Fu F L, Bucciarelli B, et al. The Medicago sativa gene index 1.2: A web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genomics, 2015, 16(1): 502. |
| 36 | Liu H, Li X Y, Zi Y F, et al. Characterization of the heat shock transcription factor family in Medicago sativa L. and its potential roles in response to abiotic stresses. International Journal of Molecular Sciences, 2023, 24(16): 12683. |
| 37 | Kang C H, Jung W Y, Kang Y H, et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death and Differentiation, 2006, 13(1): 84-95. |
| 38 | Wu R H, Guo L, Guo Y Y, et al. The G2-Like gene family in Populus trichocarpa: Identification, evolution and expression profiles. BMC Genomic Data, 2023, 24(1): 37. |
| 39 | Meng X X, Liang Z K, Dai X R, et al. Predicting transcriptional responses to cold stress across plant species. PNAS, 2021, 118(10): e2026330118. |
| 40 | Xiong B, Gong Y, Li Q, et al. Genome-wide analysis of the GLK gene family and the expression under different growth stages and dark stress in sweet orange (Citrus sinensis). Horticulturae, 2022, 8(11): 1076. |
| 41 | Zhang X Y, Zhao L J, Li Y J, et al. Expression of AtPUB18 after salt stress treatment and analysis of its promoter from Arabidopsis thaliana. Acta Botanica Boreali-Occidentalia Sinica, 2014, 34(1): 54-59. |
| 张新宇, 赵兰杰, 李艳军, 等. 盐胁迫对拟南芥AtPUB18基因的诱导表达及其启动子分析. 西北植物学报, 2014, 34(1): 54-59. | |
| 42 | Panchy N, Lehti-Shiu M, Shiu S H. Evolution of gene duplication in plants. Plant Physiology, 2016, 171(4): 2294-2316. |
| 43 | Cannon S B, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology, 2004, 4: 10. |
| 44 | Birchler J A, Yang H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. The Plant Cell, 2022, 34(7): 2466-2474. |
| 45 | Ram H, Sahadevan S, Gale N, et al. An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PLoS Genetics, 2020, 16(4): e1008661. |
| 46 | Kiba T, Inaba J, Kudo T, et al. Repression of nitrogen starvation responses by members of the Arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. The Plant Cell, 2018, 30(4): 925-945. |
| 47 | Li Q, Zhou L Y, Li Y H, et al. Plant NIGT1/HRS1/HHO transcription factors: Key regulators with multiple roles in plant growth, development, and stress responses. International Journal of Molecular Sciences, 2021, 22(16): 8685. |
| 48 | Zhao X Y, Yang J J, Li X Z, et al. Identification and expression analysis of GARP superfamily genes in response to nitrogen and phosphorus stress in Spirodela polyrhiza. BMC Plant Biology, 2022, 22(1): 308. |
| 49 | Rose A, Meier I, Wienand U. The tomato I-box binding factor LeMYBI is a member of a novel class of myb-like proteins. The Plant Journal, 1999, 20(6): 641-652. |
| 50 | Rogozin I B, Carmel L, Csuros M, et al. Origin and evolution of spliceosomal introns. Biology Direct, 2012, 7: 11. |
| 51 | Yang L L, Zhang X Y, Wang L Y, et al. Lineage-specific amplification and epigenetic regulation of LTR-retrotransposons contribute to the structure, evolution, and function of Fabaceae species. BMC Genomics, 2023, 24(1): 423. |
| 52 | Fitter D W, Martin D J, Copley M J, et al. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal, 2010, 31(6): 713-727. |
| 53 | Tamai H, Iwabuchi M, Meshi T. Arabidopsis GARP transcriptional activators interact with the pro-rich activation domain shared by G-Box-Binding bZIP factors. Plant and Cell Physiology, 2002, 43(1): 99-107. |
| 54 | Lee S C, Kim S H, Kim S R. Drought inducible OsDhn1 promoter is activated by OsDREB1A and OsDREB1D. Journal of Integrative Plant Biology, 2013, 56: 115-121. |
| 55 | Wang J L, Du C X, Zhou H K, et al. Exogenic ABA and plant abiotic stress resistance mechanism. Journal of Anhui Agricultural Sciences, 2019, 47(13): 12-15. |
| 王金丽, 杜晨曦, 周华坤, 等. 外源ABA与植物非生物胁迫抗逆机制. 安徽农业科学, 2019, 47(13): 12-15. | |
| 56 | Yoshida T, Fernie A R, Shinozaki K, et al. Long-distance stress and developmental signals associated with abscisic acid signaling in environmental responses. The Plant Journal: For Cell and Molecular Biology, 2021, 105(2): 477-488. |
| [1] | 贺龙义, 谭萌萌, 车海涛, 张红鹰, 朱雨欣, 张彦妮. 细叶百合LpDREB9基因克隆及耐旱性分析[J]. 草业学报, 2025, 34(1): 161-173. |
| [2] | 王晓彤, 李小红, 麻旭霞, 蔡文祺, 冯学丽, 李淑霞. 紫花苜蓿FBA基因家族成员的鉴定与分析[J]. 草业学报, 2024, 33(9): 81-93. |
| [3] | 张盈盈, 胡丹丹, 马春晖, 张前兵. 苜蓿叶片结构和光合特性对菌磷添加的响应[J]. 草业学报, 2024, 33(8): 133-144. |
| [4] | 王峥, 常伟, 李俊诚, 苏连泰, 高鲤, 周鹏, 安渊. 紫花苜蓿还田对饲料玉米产量和氮素吸收转运的影响[J]. 草业学报, 2024, 33(8): 63-73. |
| [5] | 吴毅, 冯雅岚, 王添宁, 琚吉浩, 肖慧淑, 马超, 张均. 小麦及其祖先物种Hsp70基因家族鉴定与表达分析[J]. 草业学报, 2024, 33(7): 53-67. |
| [6] | 张震欢, 姚立蓉, 汪军成, 司二静, 张宏, 杨轲, 马小乐, 孟亚雄, 王化俊, 李葆春. 盐生草AKR基因家族成员的鉴定及根系盐胁迫响应基因HgAKR42639的耐盐分析[J]. 草业学报, 2024, 33(7): 68-83. |
| [7] | 高金柱, 赵东豪, 高乐, 苏喜浩, 何学青. 硝酸铈与脱落酸处理对紫花苜蓿种子萌发和幼苗生理特性的影响[J]. 草业学报, 2024, 33(6): 175-186. |
| [8] | 谭英, 尹豪. 盐胁迫下根施AMF和褪黑素对紫花苜蓿生长、光合特征以及抗氧化系统的影响[J]. 草业学报, 2024, 33(6): 64-75. |
| [9] | 王敏, 李莉, 贾蓉, 包爱科. 10种紫花苜蓿在低温胁迫下的生理特性及耐寒性评价[J]. 草业学报, 2024, 33(6): 76-88. |
| [10] | 孔海明, 宋家兴, 杨静, 李倩, 杨培志, 曹玉曼. 紫花苜蓿CAMTA基因家族鉴定及其在非生物胁迫下的表达模式分析[J]. 草业学报, 2024, 33(5): 143-154. |
| [11] | 何升然, 刘晓静, 赵雅姣, 汪雪, 王静. 紫花苜蓿/甜高粱间作对根际土壤特性及微生物群落特征的影响[J]. 草业学报, 2024, 33(5): 92-105. |
| [12] | 刘昊, 李显炀, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 紫花苜蓿SAUR基因家族的鉴定及其在非生物胁迫中的表达模式研究[J]. 草业学报, 2024, 33(4): 135-153. |
| [13] | 李显炀, 刘昊, 何飞, 王雪, 李明娜, 龙瑞才, 康俊梅, 杨青川, 陈林. 全基因组水平紫花苜蓿WRKY转录因子家族鉴定与表达模式分析[J]. 草业学报, 2024, 33(4): 154-170. |
| [14] | 李妍, 马富龙, 韩路, 王海珍. 美国‘WL’系列不同秋眠级苜蓿品种在南疆的生产性能与适应性评价[J]. 草业学报, 2024, 33(3): 139-149. |
| [15] | 汪雪, 刘晓静, 王静, 吴勇, 童长春. 连续间作下的紫花苜蓿/燕麦根系与碳氮代谢特性研究[J]. 草业学报, 2024, 33(3): 85-96. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||