

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (8): 145-158.DOI: 10.11686/cyxb2023346
• 研究论文 • 上一篇
马圆1( ), 刘欢1(
), 刘欢1( ), 赵桂琴1, 王敬龙2, 张然3, 姚瑞瑞1
), 赵桂琴1, 王敬龙2, 张然3, 姚瑞瑞1
收稿日期:2023-09-19
修回日期:2023-11-01
出版日期:2024-08-20
发布日期:2024-05-13
通讯作者:
刘欢
作者简介:E-mail: liuhuan@gsau.edu.cn基金资助:
Yuan MA1( ), Huan LIU1(
), Huan LIU1( ), Gui-qin ZHAO1, Jing-long WANG2, Ran ZHANG3, Rui-rui YAO1
), Gui-qin ZHAO1, Jing-long WANG2, Ran ZHANG3, Rui-rui YAO1
Received:2023-09-19
Revised:2023-11-01
Online:2024-08-20
Published:2024-05-13
Contact:
Huan LIU
摘要:
小热激蛋白(sHSPs)是植物体中普遍存在的由核基因编码的一类蛋白,具有保守的ACD结构域,能够在高温、干旱以及老化等胁迫刺激中发挥重要作用。本研究利用生物信息学方法在燕麦基因组中鉴定得到24个HSP20(AsHSP20.1~AsHSP20.24)基因,并对AsHSP20家族成员的理化性质、蛋白结构、亚细胞定位、系统进化、保守基序和保守结构域、基因染色体位置以及响应高温及老化基因的表达等进行系统分析。研究发现AsHSP20基因分布在17条染色体上。编码氨基酸数目136~529 aa,分子量大小14.9~58.1 kDa,理论等电点5.30~8.79。HSP20成员多定位在核、胞质和叶绿体上,部分定位在质膜、线粒体、过氧化物酶体以及胞外。蛋白质二级结构和三级结构分析表明AsHSP20成员确定有β-折叠结构。根据保守基序组成与系统发育关系分析,AsHSP20基因家族被分为11个亚组,同一亚组成员之间具有相似或相同的保守基序,表明这些蛋白之间具有功能的相似性。进一步分析自然老化和人工老化处理下AsHSP20基因的表达情况,结果表明AsHSP20.20、AsHSP20.24基因在自然老化处理与人工老化处理下共同下调表达,推测AsHSP20.20和AsHSP20.24在两种老化方法下共同参与调控燕麦种子活力降低的过程,可作为燕麦种子寿命及抗老化种质育种研究的候选基因。研究为AsHSP20基因家族在燕麦种质老化过程中的调控机制提供了有价值的信息,也为进一步探究燕麦HSP20基因的功能及种子抗衰老分子机制提供了理论支撑。
马圆, 刘欢, 赵桂琴, 王敬龙, 张然, 姚瑞瑞. 燕麦sHSP基因家族的鉴定及其响应高温及老化的表达分析[J]. 草业学报, 2024, 33(8): 145-158.
Yuan MA, Huan LIU, Gui-qin ZHAO, Jing-long WANG, Ran ZHANG, Rui-rui YAO. Identification of the oat sHSP gene family and its transcript profiles in response to high temperature and aging[J]. Acta Prataculturae Sinica, 2024, 33(8): 145-158.
| 名称Name | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| AsHSP20.1 | GCCGCTCAGTATTATGCAAG | CCGATCAAGGGGTTCAAGTA |
| AsHSP20.2 | TCAGCTGGAACGAATAGTCC | AGTAACGATGGAGAAGCCAG |
| AsHSP20.3 | TAGGGGACCAATCTCTGTGT | TTGGTGCATTTCAGAAGGGT |
| AsHSP20.4 | GAGCTCTTCTTCGGGAGC | ACCTTGACGTCGTCCTTG |
| AsHSP20.5 | ATCTTCCAGGGGTGAAGAAG | CCACTTGTCGTTCTTGTCCT |
| AsHSP20.6 | CATCTACTCTGCCGACCTC | TTCCTGGCGAAACTGCTC |
| AsHSP20.7 | ATCTCTACTCTGCCGACCTC | GATGACGAGGTATCTCCCG |
| AsHSP20.8 | TTTGTTGATCAGCAGAGGGT | TATGTATGGCATCCGAGACG |
| AsHSP20.9 | CTCCGATAAGCCAACGGATA | TTGCTACTCGTTGCAGTTTG |
| AsHSP20.10 | GAAGTACCTGCGGATGGA | GCTTCTCAACCAACACGGT |
| AsHSP20.11 | AAGGAGGACGCCAAGTACC | CTTCTCCATGTCGGCGTT |
| AsHSP20.12 | AAGTTCGTTAGGCGTTTTCG | GCTTCTTGACCTCGGTTTTG |
| AsHSP20.13 | ACAAGAACGAGAAATGGCAC | ATATCTCGATGGCCTTCACC |
| AsHSP20.14 | GAGCATCGACATCCTGGAG | CACCTTCACCTCCTCCTTG |
| AsHSP20.15 | TGTTCAACACCCAGGGTTC | CAGGTCCTGCGAGAAGAAG |
| AsHSP20.16 | CGCGTACAAGATGGACAAGA | CTCCTCCTTCTTGAGCTTGG |
| AsHSP20.17 | GTTCAAGTTGCCAGACAATG | ATCTGGATGGATTTCACCTCG |
| AsHSP20.18 | GTGTTTGACCCCTTCTCTCT | ACCTTCACCTCCTCCTTCTT |
| AsHSP20.19 | GACGTACCTCGACTTCATGC | TCAGGTGCTCCTTCTTGAAC |
| AsHSP20.20 | CTCTCTCGATCTCTTGGACC | TTCACCTCCTCCTTCTTGAG |
| AsHSP20.21 | AGGGATGCCAATTTCTCGAT | TTCTTGTGTGAGGTGCTCTT |
| AsHSP20.22 | GTGCATGGTTCTTCTTCACC | GAAAGCATCCATTGTGGCAT |
| AsHSP20.23 | GACATTGTGGAGGACGAGAA | CCACCATCACGTTCACCT |
| AsHSP20.24 | GTTTTTCTCGCGATGACCG | CCACCGCCTGAGAAGAGAT |
| AsActin(KP257585.1) | CATTGGTATGGAAGCTGCTG | CACTGAGCACAATGTTACCG |
表1 HSP20基因的qRT-PCR引物
Table 1 qRT-PCR primers of HSP20 genes
| 名称Name | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| AsHSP20.1 | GCCGCTCAGTATTATGCAAG | CCGATCAAGGGGTTCAAGTA |
| AsHSP20.2 | TCAGCTGGAACGAATAGTCC | AGTAACGATGGAGAAGCCAG |
| AsHSP20.3 | TAGGGGACCAATCTCTGTGT | TTGGTGCATTTCAGAAGGGT |
| AsHSP20.4 | GAGCTCTTCTTCGGGAGC | ACCTTGACGTCGTCCTTG |
| AsHSP20.5 | ATCTTCCAGGGGTGAAGAAG | CCACTTGTCGTTCTTGTCCT |
| AsHSP20.6 | CATCTACTCTGCCGACCTC | TTCCTGGCGAAACTGCTC |
| AsHSP20.7 | ATCTCTACTCTGCCGACCTC | GATGACGAGGTATCTCCCG |
| AsHSP20.8 | TTTGTTGATCAGCAGAGGGT | TATGTATGGCATCCGAGACG |
| AsHSP20.9 | CTCCGATAAGCCAACGGATA | TTGCTACTCGTTGCAGTTTG |
| AsHSP20.10 | GAAGTACCTGCGGATGGA | GCTTCTCAACCAACACGGT |
| AsHSP20.11 | AAGGAGGACGCCAAGTACC | CTTCTCCATGTCGGCGTT |
| AsHSP20.12 | AAGTTCGTTAGGCGTTTTCG | GCTTCTTGACCTCGGTTTTG |
| AsHSP20.13 | ACAAGAACGAGAAATGGCAC | ATATCTCGATGGCCTTCACC |
| AsHSP20.14 | GAGCATCGACATCCTGGAG | CACCTTCACCTCCTCCTTG |
| AsHSP20.15 | TGTTCAACACCCAGGGTTC | CAGGTCCTGCGAGAAGAAG |
| AsHSP20.16 | CGCGTACAAGATGGACAAGA | CTCCTCCTTCTTGAGCTTGG |
| AsHSP20.17 | GTTCAAGTTGCCAGACAATG | ATCTGGATGGATTTCACCTCG |
| AsHSP20.18 | GTGTTTGACCCCTTCTCTCT | ACCTTCACCTCCTCCTTCTT |
| AsHSP20.19 | GACGTACCTCGACTTCATGC | TCAGGTGCTCCTTCTTGAAC |
| AsHSP20.20 | CTCTCTCGATCTCTTGGACC | TTCACCTCCTCCTTCTTGAG |
| AsHSP20.21 | AGGGATGCCAATTTCTCGAT | TTCTTGTGTGAGGTGCTCTT |
| AsHSP20.22 | GTGCATGGTTCTTCTTCACC | GAAAGCATCCATTGTGGCAT |
| AsHSP20.23 | GACATTGTGGAGGACGAGAA | CCACCATCACGTTCACCT |
| AsHSP20.24 | GTTTTTCTCGCGATGACCG | CCACCGCCTGAGAAGAGAT |
| AsActin(KP257585.1) | CATTGGTATGGAAGCTGCTG | CACTGAGCACAATGTTACCG |
| 基因名Gene name | 重命名Rename | ORF (bp) | aa | Mw (Da) | IP | II | PH | SLP |
|---|---|---|---|---|---|---|---|---|
| AVESA.00010b.r2.6AG1028780.1 | AsHSP20.1 | 1281 | 426 | 46386.23 | 8.51 | 46.41 | -0.123 | Chl |
| AVESA.00010b.r2.6AG1028780.2 | AsHSP20.2 | 1035 | 344 | 37976.51 | 5.76 | 41.26 | -0.162 | Chl |
| AVESA.00010b.r2.6DG1171000.1 | AsHSP20.3 | 573 | 190 | 21219.33 | 6.59 | 47.41 | -0.153 | Chl |
| AVESA.00010b.r2.7AG1212080.1 | AsHSP20.4 | 411 | 136 | 14949.18 | 8.01 | 41.96 | -0.302 | Per |
| AVESA.00010b.r2.3AG0406840.2 | AsHSP20.5 | 456 | 151 | 16900.14 | 6.20 | 44.96 | -0.597 | Cyt |
| AVESA.00010b.r2.2DG0350420.1 | AsHSP20.6 | 468 | 155 | 17251.45 | 6.19 | 52.50 | -0.408 | Cyt |
| AVESA.00010b.r2.2CG0275380.1 | AsHSP20.7 | 468 | 155 | 17304.55 | 6.51 | 49.79 | -0.434 | Cyt |
| AVESA.00010b.r2.4AG0636730.1 | AsHSP20.8 | 1557 | 518 | 57622.58 | 6.63 | 50.48 | -0.459 | Pl |
| AVESA.00010b.r2.3AG0433390.1 | AsHSP20.9 | 828 | 275 | 29972.66 | 8.79 | 42.43 | -0.759 | Ex |
| AVESA.00010b.r2.7CG0689040.1 | AsHSP20.10 | 480 | 159 | 17528.28 | 5.37 | 36.19 | -0.382 | Cyt |
| AVESA.00010b.r2.3DG0518610.1 | AsHSP20.11 | 480 | 159 | 17508.25 | 5.71 | 36.38 | -0.458 | Cyt |
| AVESA.00010b.r2.3AG0406840.1 | AsHSP20.12 | 456 | 151 | 17034.29 | 5.83 | 45.89 | -0.647 | Cyt |
| AVESA.00010b.r2.3CG0454310.1 | AsHSP20.13 | 456 | 151 | 16926.12 | 6.00 | 56.17 | -0.570 | Cyt |
| AVESA.00010b.r2.6CG1118770.1 | AsHSP20.14 | 534 | 177 | 19298.95 | 6.76 | 48.07 | -0.473 | Nuc |
| AVESA.00010b.r2.7DG1398280.1 | AsHSP20.15 | 687 | 228 | 24631.75 | 5.30 | 41.45 | -0.420 | Mit |
| AVESA.00010b.r2.1CG0084310.1 | AsHSP20.16 | 678 | 225 | 24304.46 | 5.61 | 45.15 | -0.498 | Mit |
| AVESA.00010b.r2.4CG1325780.1 | AsHSP20.17 | 474 | 157 | 17411.57 | 5.80 | 54.23 | -0.628 | Nuc |
| AVESA.00010b.r2.1DG0138620.1 | AsHSP20.18 | 450 | 149 | 16732.84 | 5.36 | 48.94 | -0.714 | Nuc |
| AVESA.00010b.r2.4AG0623840.1 | AsHSP20.19 | 663 | 220 | 23889.56 | 5.41 | 47.70 | -0.556 | Ex |
| AVESA.00010b.r2.4CG1325690.1 | AsHSP20.20 | 465 | 154 | 17198.33 | 5.68 | 44.63 | -0.633 | Nuc |
| AVESA.00010b.r2.5DG1005630.2 | AsHSP20.21 | 1590 | 529 | 58060.51 | 6.29 | 48.53 | -0.474 | Pl |
| AVESA.00010b.r2.4CG1324540.2 | AsHSP20.22 | 1590 | 529 | 58067.52 | 6.21 | 50.25 | -0.465 | Pl |
| AVESA.00010b.r2.5AG0799690.1 | AsHSP20.23 | 738 | 245 | 26922.35 | 6.77 | 53.50 | -0.642 | Chl |
| AVESA.00010b.r2.7CG0688840.1 | AsHSP20.24 | 618 | 205 | 22139.24 | 7.91 | 39.45 | -0.279 | Nuc |
表2 燕麦HSP20基因家族理化性质
Table 2 Physicochemical properties of the HSP20 gene family of oats
| 基因名Gene name | 重命名Rename | ORF (bp) | aa | Mw (Da) | IP | II | PH | SLP |
|---|---|---|---|---|---|---|---|---|
| AVESA.00010b.r2.6AG1028780.1 | AsHSP20.1 | 1281 | 426 | 46386.23 | 8.51 | 46.41 | -0.123 | Chl |
| AVESA.00010b.r2.6AG1028780.2 | AsHSP20.2 | 1035 | 344 | 37976.51 | 5.76 | 41.26 | -0.162 | Chl |
| AVESA.00010b.r2.6DG1171000.1 | AsHSP20.3 | 573 | 190 | 21219.33 | 6.59 | 47.41 | -0.153 | Chl |
| AVESA.00010b.r2.7AG1212080.1 | AsHSP20.4 | 411 | 136 | 14949.18 | 8.01 | 41.96 | -0.302 | Per |
| AVESA.00010b.r2.3AG0406840.2 | AsHSP20.5 | 456 | 151 | 16900.14 | 6.20 | 44.96 | -0.597 | Cyt |
| AVESA.00010b.r2.2DG0350420.1 | AsHSP20.6 | 468 | 155 | 17251.45 | 6.19 | 52.50 | -0.408 | Cyt |
| AVESA.00010b.r2.2CG0275380.1 | AsHSP20.7 | 468 | 155 | 17304.55 | 6.51 | 49.79 | -0.434 | Cyt |
| AVESA.00010b.r2.4AG0636730.1 | AsHSP20.8 | 1557 | 518 | 57622.58 | 6.63 | 50.48 | -0.459 | Pl |
| AVESA.00010b.r2.3AG0433390.1 | AsHSP20.9 | 828 | 275 | 29972.66 | 8.79 | 42.43 | -0.759 | Ex |
| AVESA.00010b.r2.7CG0689040.1 | AsHSP20.10 | 480 | 159 | 17528.28 | 5.37 | 36.19 | -0.382 | Cyt |
| AVESA.00010b.r2.3DG0518610.1 | AsHSP20.11 | 480 | 159 | 17508.25 | 5.71 | 36.38 | -0.458 | Cyt |
| AVESA.00010b.r2.3AG0406840.1 | AsHSP20.12 | 456 | 151 | 17034.29 | 5.83 | 45.89 | -0.647 | Cyt |
| AVESA.00010b.r2.3CG0454310.1 | AsHSP20.13 | 456 | 151 | 16926.12 | 6.00 | 56.17 | -0.570 | Cyt |
| AVESA.00010b.r2.6CG1118770.1 | AsHSP20.14 | 534 | 177 | 19298.95 | 6.76 | 48.07 | -0.473 | Nuc |
| AVESA.00010b.r2.7DG1398280.1 | AsHSP20.15 | 687 | 228 | 24631.75 | 5.30 | 41.45 | -0.420 | Mit |
| AVESA.00010b.r2.1CG0084310.1 | AsHSP20.16 | 678 | 225 | 24304.46 | 5.61 | 45.15 | -0.498 | Mit |
| AVESA.00010b.r2.4CG1325780.1 | AsHSP20.17 | 474 | 157 | 17411.57 | 5.80 | 54.23 | -0.628 | Nuc |
| AVESA.00010b.r2.1DG0138620.1 | AsHSP20.18 | 450 | 149 | 16732.84 | 5.36 | 48.94 | -0.714 | Nuc |
| AVESA.00010b.r2.4AG0623840.1 | AsHSP20.19 | 663 | 220 | 23889.56 | 5.41 | 47.70 | -0.556 | Ex |
| AVESA.00010b.r2.4CG1325690.1 | AsHSP20.20 | 465 | 154 | 17198.33 | 5.68 | 44.63 | -0.633 | Nuc |
| AVESA.00010b.r2.5DG1005630.2 | AsHSP20.21 | 1590 | 529 | 58060.51 | 6.29 | 48.53 | -0.474 | Pl |
| AVESA.00010b.r2.4CG1324540.2 | AsHSP20.22 | 1590 | 529 | 58067.52 | 6.21 | 50.25 | -0.465 | Pl |
| AVESA.00010b.r2.5AG0799690.1 | AsHSP20.23 | 738 | 245 | 26922.35 | 6.77 | 53.50 | -0.642 | Chl |
| AVESA.00010b.r2.7CG0688840.1 | AsHSP20.24 | 618 | 205 | 22139.24 | 7.91 | 39.45 | -0.279 | Nuc |
名称 Name | α-螺旋 Alpha-helix | 延伸链 Extended strand | 无规则卷曲 Random coil | β-转角 Beta turn | β-折叠 Beta sheet | 名称 Name | α-螺旋 Alpha-helix | 延伸链 Extended strand | 无规则卷曲 Random coil | β-转角 Beta turn | β-折叠 Beta sheet |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AsHSP20.1 | 40.38 | 17.84 | 37.09 | 4.69 | 17 | AsHSP20.13 | 17.22 | 19.21 | 58.28 | 5.30 | 40 |
| AsHSP20.2 | 43.90 | 15.12 | 35.76 | 5.23 | 20 | AsHSP20.14 | 23.16 | 20.90 | 48.59 | 7.34 | 36 |
| AsHSP20.3 | 30.00 | 23.68 | 39.47 | 6.84 | 32 | AsHSP20.15 | 28.95 | 21.93 | 43.42 | 5.70 | 22 |
| AsHSP20.4 | 23.53 | 22.79 | 47.06 | 6.62 | 48 | AsHSP20.16 | 21.33 | 16.89 | 57.78 | 4.00 | 24 |
| AsHSP20.5 | 23.18 | 20.53 | 50.33 | 5.96 | 38 | AsHSP20.17 | 15.29 | 19.75 | 57.32 | 7.64 | 38 |
| AsHSP20.6 | 17.42 | 24.52 | 49.68 | 8.39 | 30 | AsHSP20.18 | 30.20 | 18.79 | 44.30 | 6.71 | 39 |
| AsHSP20.7 | 18.06 | 25.16 | 48.39 | 8.39 | 32 | AsHSP20.19 | 43.18 | 16.82 | 29.55 | 10.45 | 30 |
| AsHSP20.8 | 19.88 | 18.53 | 54.63 | 6.95 | 27 | AsHSP20.20 | 17.53 | 22.08 | 55.19 | 5.19 | 38 |
| AsHSP20.9 | 42.55 | 12.00 | 40.00 | 5.45 | 16 | AsHSP20.21 | 18.15 | 20.23 | 56.14 | 5.48 | 23 |
| AsHSP20.10 | 37.74 | 20.75 | 37.11 | 4.40 | 36 | AsHSP20.22 | 16.45 | 18.34 | 59.55 | 5.67 | 24 |
| AsHSP20.11 | 33.96 | 20.13 | 40.88 | 5.03 | 36 | AsHSP20.23 | 20.82 | 15.10 | 61.22 | 2.86 | 24 |
| AsHSP20.12 | 25.17 | 19.21 | 48.34 | 7.28 | 38 | AsHSP20.24 | 42.93 | 16.59 | 34.63 | 5.85 | 30 |
表3 燕麦HSP20基因家族蛋白质二级结构预测
Table 3 Prediction of secondary structure of HSP20 gene family protein in oats (%)
名称 Name | α-螺旋 Alpha-helix | 延伸链 Extended strand | 无规则卷曲 Random coil | β-转角 Beta turn | β-折叠 Beta sheet | 名称 Name | α-螺旋 Alpha-helix | 延伸链 Extended strand | 无规则卷曲 Random coil | β-转角 Beta turn | β-折叠 Beta sheet |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AsHSP20.1 | 40.38 | 17.84 | 37.09 | 4.69 | 17 | AsHSP20.13 | 17.22 | 19.21 | 58.28 | 5.30 | 40 |
| AsHSP20.2 | 43.90 | 15.12 | 35.76 | 5.23 | 20 | AsHSP20.14 | 23.16 | 20.90 | 48.59 | 7.34 | 36 |
| AsHSP20.3 | 30.00 | 23.68 | 39.47 | 6.84 | 32 | AsHSP20.15 | 28.95 | 21.93 | 43.42 | 5.70 | 22 |
| AsHSP20.4 | 23.53 | 22.79 | 47.06 | 6.62 | 48 | AsHSP20.16 | 21.33 | 16.89 | 57.78 | 4.00 | 24 |
| AsHSP20.5 | 23.18 | 20.53 | 50.33 | 5.96 | 38 | AsHSP20.17 | 15.29 | 19.75 | 57.32 | 7.64 | 38 |
| AsHSP20.6 | 17.42 | 24.52 | 49.68 | 8.39 | 30 | AsHSP20.18 | 30.20 | 18.79 | 44.30 | 6.71 | 39 |
| AsHSP20.7 | 18.06 | 25.16 | 48.39 | 8.39 | 32 | AsHSP20.19 | 43.18 | 16.82 | 29.55 | 10.45 | 30 |
| AsHSP20.8 | 19.88 | 18.53 | 54.63 | 6.95 | 27 | AsHSP20.20 | 17.53 | 22.08 | 55.19 | 5.19 | 38 |
| AsHSP20.9 | 42.55 | 12.00 | 40.00 | 5.45 | 16 | AsHSP20.21 | 18.15 | 20.23 | 56.14 | 5.48 | 23 |
| AsHSP20.10 | 37.74 | 20.75 | 37.11 | 4.40 | 36 | AsHSP20.22 | 16.45 | 18.34 | 59.55 | 5.67 | 24 |
| AsHSP20.11 | 33.96 | 20.13 | 40.88 | 5.03 | 36 | AsHSP20.23 | 20.82 | 15.10 | 61.22 | 2.86 | 24 |
| AsHSP20.12 | 25.17 | 19.21 | 48.34 | 7.28 | 38 | AsHSP20.24 | 42.93 | 16.59 | 34.63 | 5.85 | 30 |
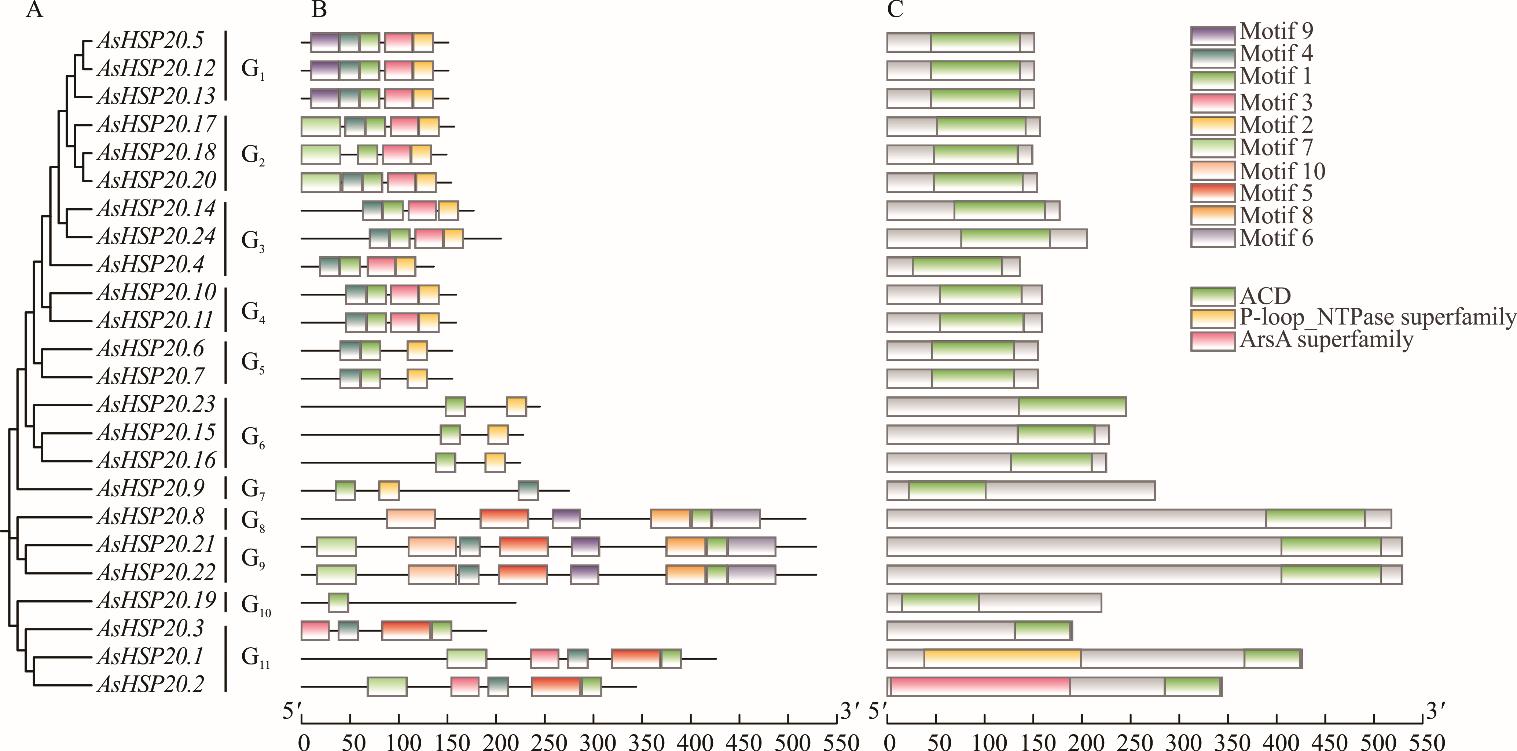
图3 燕麦HSP20基因家族系统发育树、保守基序及保守结构域A: 燕麦HSP20基因家族系统发育树Phylogenetic tree of the HSP20 gene family of oats; B: 燕麦HSP20基因家族保守基序Oat HSP20 gene family conserves motif order; C: 燕麦HSP20基因家族保守结构域The oat HSP20 gene family conserves domain.
Fig.3 Phylogenetic tree, conserved motif and conserved domain of oat HSP20 gene family
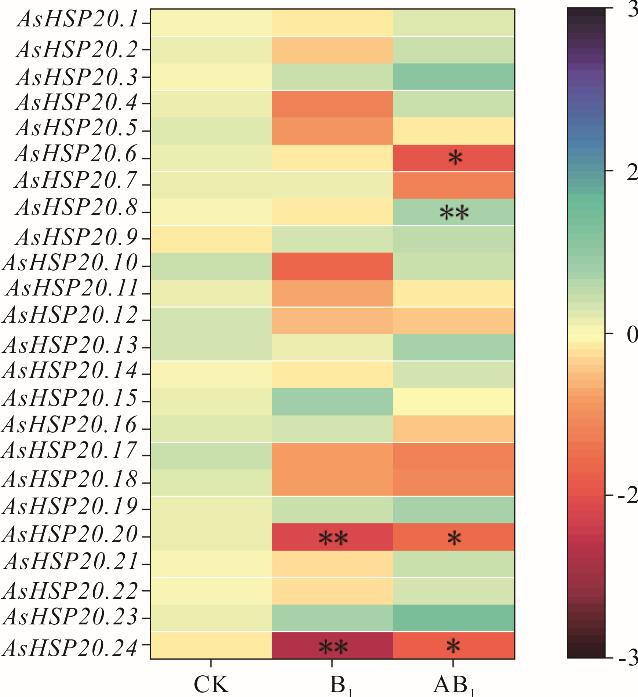
图5 AsHSP20在自然老化与人工老化下相对表达情况CK:未经过处理的种子Untreated seeds;B1:自然贮藏4年的种子Seeds stored naturally for 4 years;AB1:人工老化72 h的种子Seeds artificially aged for 72 hours; *: P<0.05; **: P<0.01; 右栏为基因表达量The right column shows gene expression.
Fig.5 Relative expression of AsHSP20 under natural aging and artificial aging
| 1 | Guo H X, Wang C Y, Zhao L, et al. Research progress of small heat shock protein. Journal of Shanxi Agricultural Sciences, 2013, 41(12): 1421-1423. |
| 郭虹霞, 王创云, 赵丽, 等. 小分子热激蛋白的研究进展. 山西农业科学, 2013, 41(12): 1421-1423. | |
| 2 | Zhang N, Jiang J. Research advances of small heat shock protein gene family (sHSPs) in plants. Plant Physiology Journal, 2017, 53(6): 943-948. |
| 张宁, 姜晶. 植物中小分子热激蛋白基因家族(sHSPs)研究进展. 植物生理学报, 2017, 53(6): 943-948. | |
| 3 | Zhao P, Wang D D, Wang R Q, et al. Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics, 2018, 19(1): 1-13. |
| 4 | Wang M, Jiang B, Lin Y E, et al. Advances of small heat shock proteins participating in plant resistance. Journal of Anhui Agricultural Sciences, 2018, 46(18): 29-32, 35. |
| 王敏, 江彪, 林毓娥, 等. 小分子热激蛋白参与植物抗逆性方面的研究进展. 安徽农业科学, 2018, 46(18): 29-32, 35. | |
| 5 | Wang T, Tang S H, He M, et al. Advances in relationship between heat shock proteins and thermo-tolerance of plants. Journal of Northwest Forestry University, 2014, 29(6): 72-79. |
| 王涛, 唐颂豪, 何梅, 等. 热激蛋白与植物耐热性关系的研究进展. 西北林学院学报, 2014, 29(6): 72-79. | |
| 6 | Zhou L, He F, Pan Q H, et al. Role of heat shock proteins in plant response to environmental stresses. Journal of Tropical Biology, 2011, 2(4): 297-301. |
| 周丽, 何非, 潘秋红, 等. 热休克蛋白在植物抗逆反应中的作用. 热带生物学报, 2011, 2(4): 297-301. | |
| 7 | Yu J H, Feng K, Cheng Y, et al. Research progress on small heat shock proteins in plants. Molecular Plant Breeding, 2017, 15(8): 3016-3023. |
| 俞佳虹, 冯坤, 程远, 等. 植物小热激蛋白的研究进展. 分子植物育种, 2017, 15(8): 3016-3023. | |
| 8 | Lubaretz O, Zur Nieden U. Accumulation of plant small heat-stress proteins in storage organs. Planta, 2002, 215(2): 220-228. |
| 9 | Al-Whaibi M H. Plant heat-shock proteins: A mini review. Journal of King Saud University-Science, 2011, 23(2): 139-150. |
| 10 | Scharf K D, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress & Chaperones, 2001, 6(3): 225-237. |
| 11 | Zhang W C, Zhu J, Yi L F. Genome-wide identification and bioinformatics analysis of HSP20 gene family in Pyropia haitanensis. Journal of Southern Agriculture, 2021, 52(6): 1527-1535. |
| 张宛晨, 朱静, 易乐飞. 坛紫菜HSP20基因家族成员的全基因组鉴定及生物信息学分析. 南方农业学报, 2021, 52(6): 1527-1535. | |
| 12 | Sun A Q, Ge S J, Dong W, et al. Cloning and function analysis of small heat shock protein gene ZmHSP17.7 from maize. Acta Agronomica Sinica, 2015, 41(3): 414-421. |
| 孙爱清, 葛淑娟, 董伟, 等. 玉米小分子热激蛋白ZmHSP17.7基因的克隆与功能分析. 作物学报, 2015, 41(3): 414-421. | |
| 13 | Chauhan H, Khurana N, Nijhavan A, et al. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell & Environment, 2012, 35(11): 1912-1931. |
| 14 | Jeevan K S P, Rajendra P S, Banerjee R, et al. Seed birth to death: dual functions of reactive oxygen species in seed physiology. Annals of Botany, 2015, 116(4): 663-668. |
| 15 | Reiter R J, Tan D X, Rosales-Corral S, et al. Mitochondria: central organelles for melatonin’s antioxidant and anti-aging actions. Molecules, 2018, 23(2): 509. |
| 16 | Tian Q, Xin X, Lu X X, et al. Research progress on plant seed aging and mitochondria. Journal of Plant Genetic Resources, 2012, 13(2): 283-287. |
| 田茜, 辛霞, 卢新雄, 等. 植物种子衰老与线粒体关系的研究进展. 植物遗传资源学报, 2012, 13(2): 283-287. | |
| 17 | Kong L Q. Physiological changes, protein and related antioxidant gene expression responses of Avena sativa L. seed under different aging treatment. Beijing: China Agricultural University, 2015. |
| 孔令琪. 不同老化处理对燕麦种子生理、蛋白质及抗氧化基因的影响. 北京: 中国农业大学, 2015. | |
| 18 | Huang Y, Liu H, Zhao G Q, et al. Effects of natural aging and artificial aging on germination characteristics and genetic integrity of oat seeds. Acta Agrestia Sinica, 2022, 30(8): 2066-2074. |
| 黄颖, 刘欢, 赵桂琴, 等. 自然老化和人工老化对燕麦种子萌发特性及遗传完整性的影响. 草地学报, 2022, 30(8): 2066-2074. | |
| 19 | Huang S Z. Heat shock protein synthesis in aged cabbage seeds. Plant Physiology Journal, 2000(1): 8-11. |
| 黄上志. 人工老化处理的卷心菜种子的热激蛋白合成. 植物生理学报, 2000(1): 8-11. | |
| 20 | Lin X D, Fu J R, Huang S Z. Synthesis of heat shock proteins in developing maize embryo. Journal of Plant Physiology and Molecular Biology, 2004, 30(2): 161-166. |
| 林晓东, 傅家瑞, 黄上志. 玉米胚发育过程中热激蛋白的合成. 植物生理与分子生物学学报, 2004, 30(2): 161-166. | |
| 21 | Gao J D. Study on the physiological characteristics and proteomics of seed storability of hybrid rice. Changsha: Hunan Agricultural University, 2012. |
| 高家东. 杂交水稻种子耐贮藏生理基础和蛋白质组学研究. 长沙: 湖南农业大学, 2012. | |
| 22 | Du Z. Overview of current status of oat utilization in China. Anhui Agricultural Science Bulletin, 2018, 24(20): 54-57. |
| 杜忠. 燕麦在中国的利用现状综述. 安徽农学通报, 2018, 24(20): 54-57. | |
| 23 | Siddique M, Gernhard S, von Koskull-Döring P, et al. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress and Chaperones, 2008, 13(2): 183-197. |
| 24 | Yu J H. Genome-wide identification and expression profiling of the SlHsp20 gene family in tomato. Jinhua: Zhejiang Normal University, 2017. |
| 俞佳虹. 番茄小热激蛋白SlHsp20基因家族的全基因组鉴定及表达分析. 金华: 浙江师范大学, 2017. | |
| 25 | Wang J, Gao X, Dong J, et al. Over-expression of the heat-responsive wheat gene TaHSP23.9 in transgenic Arabidopsis conferred tolerance to heat and salt stress. Frontiers in Plant Science, 2020, DOI: 10.3389/fpls.2020.00243. |
| 26 | Sarkar N K, Kim Y K, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics, 2009, DOI: 10.1186/1471-2164-10-393. |
| 27 | Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: a better web interface. Nucleic Acids Research, 2008, DOI: 10.1093/nar/gkn201. |
| 28 | Marchler-Bauer A, Derbyshire M K, Gonzales N R, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research, 2015, 43(D1): D222-D226. |
| 29 | Wilkins M R, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods in Molecular Biology, 1999, DOI: 10.1385/1-59259-584-7: 531. |
| 30 | He X J, Zhang L, Cheng Y D, et al. Cloning and expression analysis of tobacco NtPP2C37-like gene. Molecular Plant Breeding, 2019, 17(15): 4973-4977. |
| 何晓健, 张柳, 程亚东, 等. 烟草NtPP2C37-like基因的克隆及表达分析. 分子植物育种, 2019, 17(15): 4973-4977. | |
| 31 | Duan W K, Niu J B, Sun X C, et al. The MDAR gene family in pepper: identification, phylogeny and expression analyses against abiotic stress. Molecular Plant Breeding,[2023-10-25]. http://kns.cnki.net/kcms/detail/46.1068.S.20221215.1741.018.html. |
| 段伟科, 牛佳斌, 孙小川, 等. 辣椒MDAR基因鉴定、系统进化及非生物胁迫响应. 分子植物育种, [2023-10-25]. http://kns.cnki.net/kcms/detail/46.1068.S.20221215.1741.018.html. | |
| 32 | Montgomerie S, Cruz J A, Shrivastava S, et al. PROTEUS2: a web server for comprehensive protein structure prediction and structure-based annotation. Nucleic Acids Research, 2008, DOI: 10.1093/nar/gkn255. |
| 33 | Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. |
| 34 | Zhao Z Y, Wang J, Liu L P, et al. Identification and expression analysis of UBP gene family in common tobacco. Molecular Plant Breeding, [2023-10-25]. http://kns.cnki.net/kcms/detail/46.1068.s.20221130.1646.014.html. |
| 赵泽玉, 王剑, 刘利平, 等. 普通烟草UBP基因家族成员鉴定与表达分析. 分子植物育种, [2023-10-25]. http://kns.cnki.net/kcms/detail/46.1068.s.20221130.1646.014.html. | |
| 35 | Bailey T L, Boden M, Buske F A, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research, 2009, DOI: 10.1093/nar/gkp335. |
| 36 | Yang M Z, Derbyshire M K, Yamashita R A, et al. NCBI’s conserved domain database and tools for protein domain analysis. Current Protocols in Bioinformatics, 2020, 69(1): e90. |
| 37 | Chen C J, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8): 1194-1202. |
| 38 | Zheng X Y, Ren Z S, Zheng G H. Discussion on methods for determining seed viability——Ⅲ. Artificial accelerated aging method. Seed, 1982(4): 31-34. |
| 郑晓鹰, 任祝三, 郑光华. 测定种子活力方法之探讨——Ⅲ.人工加速老化法. 种子, 1982(4): 31-34. | |
| 39 | Zhu R T, Niu K J, Zhang R, et al. Identification of NAC gene family and analysis of its expression pattern under abiotic stress in Poa pratensis. Grassland and Turf, 2021, 41(4): 26-35. |
| 朱瑞婷, 牛奎举, 张然, 等. 草地早熟禾NAC基因鉴定及非生物胁迫下表达模式分析. 草原与草坪, 2021, 41(4): 26-35. | |
| 40 | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25(4): 402-408. |
| 41 | Kamal N, Tsardakas R N, Bentzer J, et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature, 2022, 606(7912): 113-119. |
| 42 | Prieto-Dapena P, Castaño R, Almoguera C, et al. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiology, 2006, 142(3): 1102-1112. |
| 43 | Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiology, 2000, 122(4): 1099-1108. |
| 44 | Tejedor-Cano J, Prieto-Dapena P, Almoguera C, et al. Loss of function of the HSFA9 seed longevity program. Plant Cell Environment, 2010, 33(8): 1408-1417. |
| 45 | Zhou W G, Meng Y J, Chen F, et al. Advances in studies of physiological and molecular mechanisms of plant seed longevity. Acta Botanica Boreali-Occidentalia Sinica, 2017, 37(2): 408-418. |
| 周文冠, 孟永杰, 陈锋, 等. 植物种子寿命的生理及分子机制研究进展. 西北植物学报, 2017, 37(2): 408-418. | |
| 46 | Zhou Y L. Function analysis of Nelumbo nucifera metallothionein and heat shock protein genes in seed vigor. Guangzhou: Sun Yat-sen University, 2011. |
| 周玉亮. 莲金属硫蛋白和热激蛋白基因在种子活力中的功能分析. 广州: 中山大学, 2011. | |
| 47 | Huang S P, Huang D Y, Fu L G, et al. Characterization of Arabidopsis thaliana Hsp20 gene family and its expression analysis under drought and salt stress. Life Science Research, 2023, 27(2): 162-169. |
| 黄思沛, 黄德娅, 付连郭, 等. 拟南芥Hsp20基因家族的特征及其在干旱和盐胁迫下的表达分析. 生命科学研究, 2023, 27(2): 162-169. | |
| 48 | Ouyang Y D, Chen J J, Xie W B, et al. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Molecular Biology, 2009, 70(3): 341-357. |
| 49 | Pandey B, Kaur A, Gupta O P, et al. Identification of HSP20 gene family in wheat and barley and their differential expression profiling under heat stress. Applied Biochemistry and Biotechnology, 2015, 175(5): 2427-2446. |
| 50 | Li J, Liu X H. Genome-wide identification and expression profile analysis of the Hsp20 gene family in barley (Hordeum vulgare L.). PeerJ, 2019, DOI: 10.7717/peerj.6832. |
| 51 | Zhang N, Jiang J, Shi J W. Genome-wide identification, phyletic evolution and expression analysis of the HSP20 gene family in tomato. Journal of Shenyang Agricultural University, 2017, 48(2): 137-144. |
| 张宁, 姜晶, 史洁玮. 番茄HSP20基因家族的全基因组鉴定、系统进化及表达分析. 沈阳农业大学学报, 2017, 48(2): 137-144. | |
| 52 | Lopes-Caitar V S, de Carvalho M C C G, Darben L M, et al. Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics, 2013, 14(1): 1-17. |
| 53 | Peng Y Y, Yan H H, Guo L C, et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nature Genetics, 2022, 54(8): 1248-1258. |
| 54 | Yu J H, Cheng Y, Feng K, et al. Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Frontiers in Plant Science, 2016, DOI: 10.3389/fpls.2016.01215. |
| 55 | Zeng C T, Qiu X W, Li D, et al. Bioinformatics analysis of the OSCA gene family in Vigna unguiculata (L.) Walp. [2023-08-28]. http://kns.cnki.net/kcms/detail/46.1068.S.20230216.0841.002.html. |
| 曾春涛, 仇学文, 李丹, 等. 豇豆OSCA基因家族生物信息学分析. [2023-08-28]. http://kns.cnki.net/kcms/detail/46.1068.S.20230216.0841.002.html. | |
| 56 | Cannon S B, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology, 2004, 4(1): 1-21. |
| 57 | Kim K H, Alam I, Kim Y G, et al. Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnology Letters, 2012, 34(2): 371-377. |
| 58 | Kaur H, Petla B P, Kamble N U, et al. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Frontiers in Plant Science, 2015, DOI: 10.3389/fpls.2015.00713. |
| 59 | Xing L M, Lv W Z, Lei W, et al. Response of HSP20 genes to artificial aging treatment in maize embryo. Acta Agronomica Sinica, 2018, 44(11): 1733-1742. |
| 邢芦蔓, 吕伟增, 雷薇, 等. 玉米种胚HSP20基因对人工老化处理的响应. 作物学报, 2018, 44(11): 1733-1742. | |
| 60 | Nagaraju M, Reddy P S, Kumar S A, et al. Genome-wide identification and transcriptional profiling of small heat shock protein gene family under diverse abiotic stress conditions in Sorghum bicolor (L.). International Journal of Biological Macromolecules, 2020, DOI: 10.1016/j.ijbiomac.2019.10.023. |
| 61 | Avelange-Macherel M H, Rolland A, Hinault M P, et al. The mitochondrial small heat shock protein HSP22 from pea is a thermosoluble chaperone prone to co-precipitate with unfolding client proteins. International Journal of Molecular Sciences, 2019, DOI: 10.3390/ijms21010097. |
| 62 | Jin X W. Effects of storage years on seed physiological and biochemical characteristics of oat. Lanzhou: Gansu Agricultural University, 2019. |
| 金小雯. 贮藏年限对燕麦种子生理生化特性的影响. 兰州: 甘肃农业大学, 2019. | |
| 63 | Liu B, Song Y M, Sun M, et al. Physiological responses of mitochondrial AsA-GSH cycle to imbibition of deteriated oat seeds. Acta Agrestia Sinica, 2021, 29(2): 211-219. |
| 刘备, 宋玉梅, 孙铭, 等. 燕麦劣变种子吸胀过程中线粒体AsA-GSH循环的生理响应. 草地学报, 2021, 29(2): 211-219. | |
| 64 | Rong J, Wang P W, Wu N, et al. Identification of heat tolerance of small heat shock protein gene HSP17.4 from soybean. Journal of Jilin Agricultural University, 2018, 40(5): 568-576. |
| 戎洁, 王丕武, 吴楠, 等. 大豆热激蛋白基因HSP17.4的耐热功能鉴定. 吉林农业大学学报, 2018, 40(5): 568-576. | |
| 65 | Rajjou L, Lovigny Y, Groot S P C, et al. Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiology, 2008, 148(1): 620-641. |
| 66 | Huang J J, Hai Z X, Wang R Y, et al. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber (Cucumis sativus L.). Frontiers in Plant Science, 2022, 13: 968418. |
| 67 | Li Y L, Li Y Q, Liu Y C, et al. The sHSP22 heat shock protein requires the ABI1 protein phosphatase to modulate polar auxin transport and downstream responses. Plant Physiology, 2018, 176(3): 2406-2425. |
| [1] | 张昭, 伏莹莹, 孙浩文, 孙逢雪, 闫慧芳. 不同品种燕麦种子活力鉴定与耐贮藏性评价[J]. 草业学报, 2024, 33(6): 165-174. |
| [2] | 李鸿飞, 周帮伟, 张淼, 施树楠, 李志坚. 不同燕麦品种在呼伦贝尔地区的引种适应性评价[J]. 草业学报, 2024, 33(4): 60-72. |
| [3] | 慕平, 柴继宽, 苏玮娟, 章海龙, 赵桂琴. 燕麦不同组合正、反交杂种后代的表型及遗传参数分析[J]. 草业学报, 2024, 33(4): 73-86. |
| [4] | 冯琴, 何小莉, 王斌, 王腾飞, 倪旺, 马霞, 明雪花, 邓建强, 兰剑. 宁夏引黄灌区燕麦与箭筈豌豆的混播效果研究[J]. 草业学报, 2024, 33(3): 107-119. |
| [5] | 鲍根生, 李媛, 冯晓云, 张鹏, 孟思宇. 高寒区氮添加和间作种植互作对燕麦和豌豆根系构型影响的研究[J]. 草业学报, 2024, 33(3): 73-84. |
| [6] | 汪雪, 刘晓静, 王静, 吴勇, 童长春. 连续间作下的紫花苜蓿/燕麦根系与碳氮代谢特性研究[J]. 草业学报, 2024, 33(3): 85-96. |
| [7] | 罗颖, 李聪, 王沛, 田莉华, 汪辉, 周青平, 雷映霞. 低氮胁迫下不同皮燕麦品种早期的响应研究及耐低氮性综合评价[J]. 草业学报, 2024, 33(2): 164-184. |
| [8] | 李文龙, 李峰, 张仲鹃, 王殿清, 王欢, 靳慧卿, 特木热, 胡志玲, 陶雅. 鄂尔多斯高原北部一年两季燕麦种植模式生产性能评价[J]. 草业学报, 2024, 33(1): 159-168. |
| [9] | 张珈敏, 关皓, 李海萍, 贾志锋, 马祥, 刘文辉, 陈有军, 陈仕勇, 蒋永梅, 甘丽, 周青平, 杨丽雪. 混播比例及乳酸菌剂对燕麦-饲用豌豆发酵TMR品质及瘤胃降解特性的影响[J]. 草业学报, 2024, 33(1): 169-181. |
| [10] | 任春燕, 梁国玲, 刘文辉, 刘凯强, 段嘉蕾. 青藏高原高寒地区早熟燕麦资源筛选和适应性评价[J]. 草业学报, 2023, 32(9): 116-129. |
| [11] | 石永红, 高鹏, 方志红, 赵祥, 韩伟, 魏江铭, 刘琳, 李锦臻. 15个进口饲用燕麦品种炭疽病的抗病性评价及损失分析[J]. 草业学报, 2023, 32(9): 130-142. |
| [12] | 者玉琦, 武志娟, 王吉坤, 钟金城, 柴志欣, 信金伟. 基于mtDNA COX3基因对西藏特色牦牛群体遗传结构的分析[J]. 草业学报, 2023, 32(9): 231-240. |
| [13] | 孙守江, 毛培胜, 豆丽茹, 贾志程, 孙铭, 马馼, 欧成明, 王娟. 活性氧及染色体端粒调控种子老化研究[J]. 草业学报, 2023, 32(8): 202-213. |
| [14] | 张振粉, 黄荣, 李向阳, 姚博, 赵桂琴. 基于Illumina MiSeq高通量测序的燕麦种带细菌多样性及功能分析[J]. 草业学报, 2023, 32(7): 96-108. |
| [15] | 王梓凡, 张晓庆, 钟志明, 权欣. 燕麦草捆和草块对彭波半细毛羊采食行为及生产性能的影响[J]. 草业学报, 2023, 32(5): 171-179. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||