

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2023, Vol. 32 ›› Issue (1): 178-191.DOI: 10.11686/cyxb2022008
• 研究论文 • 上一篇
刘福1,2( ), 陈诚1,2, 张凯旋2, 周美亮2(
), 陈诚1,2, 张凯旋2, 周美亮2( ), 张新全1(
), 张新全1( )
)
收稿日期:2022-01-06
修回日期:2022-02-24
出版日期:2023-01-20
发布日期:2022-11-07
通讯作者:
周美亮,张新全
作者简介:E-mail: zhangxq@sicau.edu.cn基金资助:
Fu LIU1,2( ), Cheng CHEN1,2, Kai-xuan ZHANG2, Mei-liang ZHOU2(
), Cheng CHEN1,2, Kai-xuan ZHANG2, Mei-liang ZHOU2( ), Xin-quan ZHANG1(
), Xin-quan ZHANG1( )
)
Received:2022-01-06
Revised:2022-02-24
Online:2023-01-20
Published:2022-11-07
Contact:
Mei-liang ZHOU,Xin-quan ZHANG
摘要:
干旱是影响植物生长发育的重要环境因素。本研究分析了日本百脉根抗旱相关基因LjbHLH34的耐旱功能,初步解析其响应干旱胁迫的分子机制,以期为百脉根抗旱分子育种提供理论基础。本研究克隆得到的LjbHLH34基因大小为711 bp、编码236个氨基酸,属bHLH转录因子家族成员。系统进化树分析显示,LjbHLH34蛋白与拟南芥bHLH Ⅳ亚家族中AtbHLH34和AtbHLH104亲缘关系较近。实时荧光定量分析表明LjbHLH34在日本百脉根的根中表达量最高,叶中次之,茎中最少,暗示其在日本百脉根多个组织中发挥作用;同时LjbHLH34基因也受聚乙二醇(PEG)和脱落酸(ABA)诱导表达。在酵母中检测发现LjbHLH34具有转录激活活性;亚细胞定位试验表明LjbHLH34蛋白定位于细胞核中。将LjbHLH34基因转入拟南芥获得过表达株系。在200 mmol·L-1甘露醇胁迫下,LjbHLH34转基因拟南芥的根长明显长于野生型。干旱处理后,野生型拟南芥比转基因拟南芥萎蔫程度更加明显,而转基因株系的相对含水量和超氧化物歧化酶(SOD)活性显著高于野生型,丙二醛(MDA)积累显著低于野生型。qRT-PCR检测发现在干旱处理之后,与抗逆相关的基因AtCAT1、AtCAT3和AtRD22在转基因株系中的表达量显著升高。上述结果表明LjbHLH34正调控植物的抗旱性。
刘福, 陈诚, 张凯旋, 周美亮, 张新全. 日本百脉根LjbHLH34基因克隆及耐旱功能鉴定[J]. 草业学报, 2023, 32(1): 178-191.
Fu LIU, Cheng CHEN, Kai-xuan ZHANG, Mei-liang ZHOU, Xin-quan ZHANG. Cloning and identification of drought tolerance function of the LjbHLH34 gene in Lotus japonicus[J]. Acta Prataculturae Sinica, 2023, 32(1): 178-191.
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) |
|---|---|
| T-LjbHLH34-F | ATGGTTTCCGCGGAAAACACC |
| T-LjbHLH34-R | TTAGGCAGCTGGTGGTCGGAG |
| M13-F | TGTAAAACGACGGCCAGT |
| pCAMBIA1307-LjbHLH34-F | |
| pCAMBIA1307-LjbHLH34-R | |
| pCAMBIA1307-F | AGGAAGTTCATTTCATTTGGA |
| pAN580-LjbHLH34-F | |
| pAN580-LjbHLH34-R | |
| PAN580-F | ATGACGCACAATCCCACTATCC |
| pGBKT7-LjbHLH34-F | |
| pGBKT7-LjbHLH34-R | |
| T7-F | TAATACGACTCACTATAGG |
| qLj-actin3-F | GTATTGTTGGCCGACCTCGT |
| qLj-actin3-R | AGCCTCAGTTAGAAGCACCG |
| qLj-bHLH34-F | ATGCAGTTCGAGTGGTGACG |
| qLj-bHLH34-R | AGACGGCAAAAATTGCCACA |
| qAt-actin7-F | TCCATGAAACAACTTACAACTCCATCA |
| qAt-actin7-R | CATCGTACTCACTCTTTGAAATCCACA |
| qAtCAT1-F | GAGATCCCCGTGGTTTTGCT |
| qAtCAT1-R | TGTGCAAACTCTCTGGGTGG |
| qAtCAT3-F | AGCTTCCAGTCAATGCTCCC |
| qAtCAT3-R | GTGAGACGTGGCTCCGATAG |
| qAtRD22-F | TTCGTCTTCCTCTGATCTGTCTTC |
| qAtRD22-R | TTTACTCCGCCTTTACCTACTTGG |
表1 引物序列
Table 1 Primer list
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) |
|---|---|
| T-LjbHLH34-F | ATGGTTTCCGCGGAAAACACC |
| T-LjbHLH34-R | TTAGGCAGCTGGTGGTCGGAG |
| M13-F | TGTAAAACGACGGCCAGT |
| pCAMBIA1307-LjbHLH34-F | |
| pCAMBIA1307-LjbHLH34-R | |
| pCAMBIA1307-F | AGGAAGTTCATTTCATTTGGA |
| pAN580-LjbHLH34-F | |
| pAN580-LjbHLH34-R | |
| PAN580-F | ATGACGCACAATCCCACTATCC |
| pGBKT7-LjbHLH34-F | |
| pGBKT7-LjbHLH34-R | |
| T7-F | TAATACGACTCACTATAGG |
| qLj-actin3-F | GTATTGTTGGCCGACCTCGT |
| qLj-actin3-R | AGCCTCAGTTAGAAGCACCG |
| qLj-bHLH34-F | ATGCAGTTCGAGTGGTGACG |
| qLj-bHLH34-R | AGACGGCAAAAATTGCCACA |
| qAt-actin7-F | TCCATGAAACAACTTACAACTCCATCA |
| qAt-actin7-R | CATCGTACTCACTCTTTGAAATCCACA |
| qAtCAT1-F | GAGATCCCCGTGGTTTTGCT |
| qAtCAT1-R | TGTGCAAACTCTCTGGGTGG |
| qAtCAT3-F | AGCTTCCAGTCAATGCTCCC |
| qAtCAT3-R | GTGAGACGTGGCTCCGATAG |
| qAtRD22-F | TTCGTCTTCCTCTGATCTGTCTTC |
| qAtRD22-R | TTTACTCCGCCTTTACCTACTTGG |
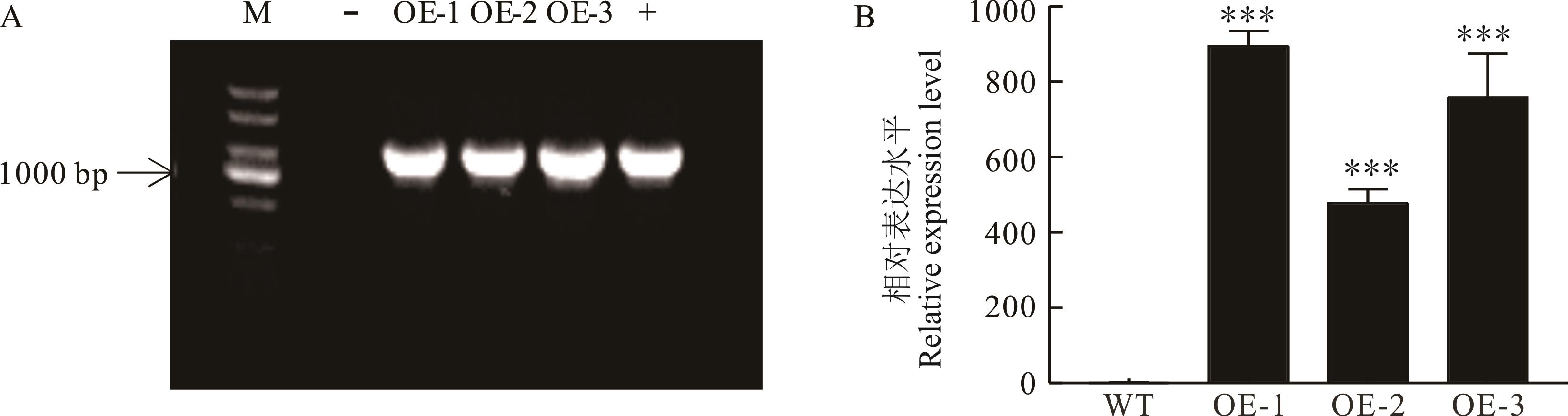
图5 转LjbHLH34基因拟南芥的鉴定M: DNA分子量标准DNA marker; -: 阴性对照Negative control; +: 阳性对照Positive control; ***表示转基因拟南芥与WT相比差异极显著(P<0.001),WT、OE-1、OE-2和OE-3分别为野生型、过表达株系1、过表达株系2和过表达株系3, 下同。*** indicates that the difference among transgenic A. thaliana and WT is extremely significant (P<0.001), WT, OE-1, OE-2 and OE-3 were wild type, overexpressed line 1, overexpressed line 2 and overexpressed line 3, the same below.
Fig.5 Identification of transgenic A. thaliana with LjbHLH34
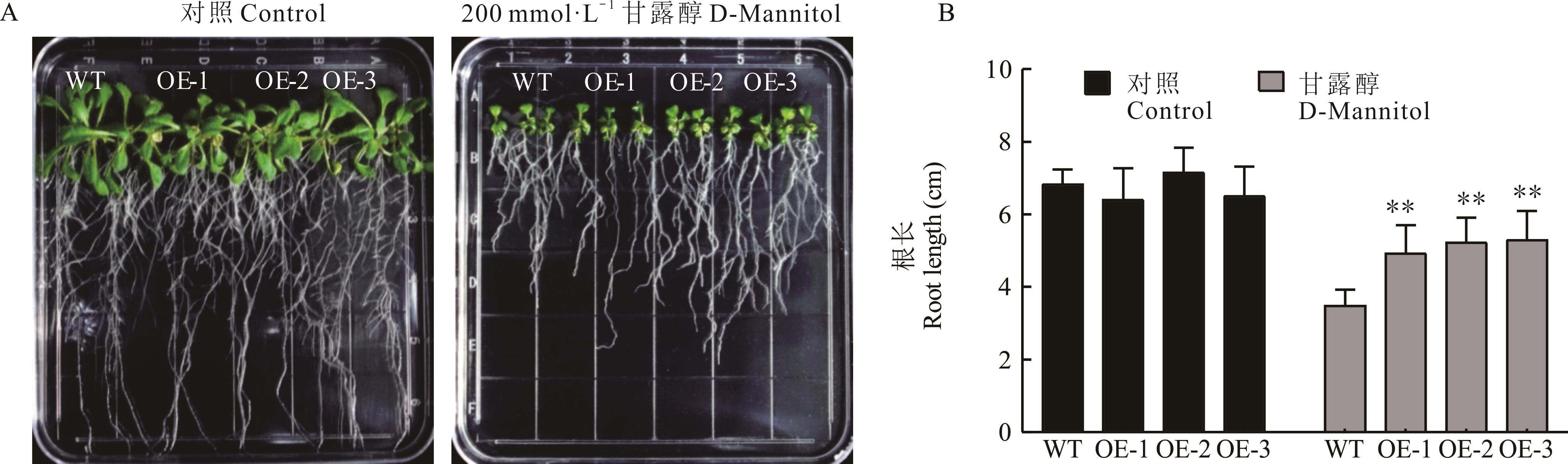
图6 甘露醇胁迫下拟南芥的根长**表示转基因拟南芥与WT相比差异显著(P<0.01)。** indicates that the difference among transgenic A. thaliana and WT is significant (P<0.01). 下同The same below.
Fig.6 Root length of A. thaliana under D-Mannitol stress
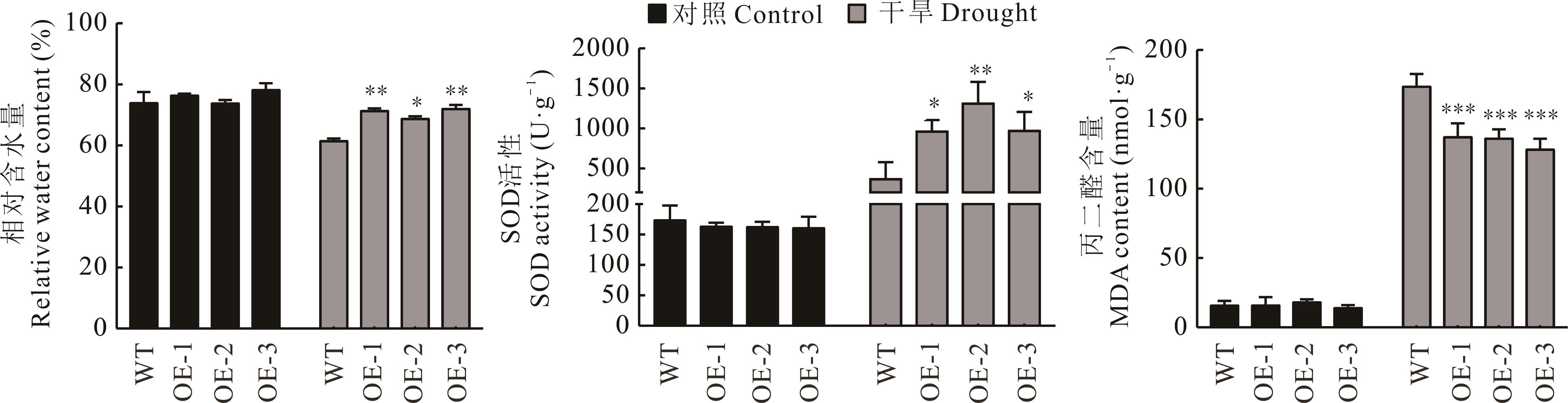
图8 干旱胁迫下转基因拟南芥的生理指标*表示转基因拟南芥与WT相比差异显著(P<0.05)。* indicates that the difference among transgenic A. thaliana and WT is significant (P<0.05). 下同The same below.
Fig.8 Physiological indexes of transgenic A. thaliana under drought stress
| 1 | Szczyglowski K, Stougaard J. Lotus genome: Pod of gold for legume research. Trends in Plant Science, 2008, 13(10): 515-517. |
| 2 | Choi H K, Mun J H, Kim D J, et al. Estimating genome conservation between crop and model legume species. Proceedings of the National Academy of Sciences, 2004, 101(43): 15289-15294. |
| 3 | Calzadilla P I, Signorelli S, Escaray F J, et al. Photosynthetic responses mediate the adaptation of two Lotus japonicus ecotypes to low temperature. Plant Science, 2016, 250: 59-68. |
| 4 | Signorelli S, Corpas F J, Borsani O, et al. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant Science, 2013, 201/202: 137-146. |
| 5 | Díaz P, Monza J, Márquez A. Drought and saline stress. Springer Netherlands, 2005, DOI: 10.1007/1-4020-3735-X_3. |
| 6 | Gong Z Z, Xiong L M, Shi H Z, et al. Plant abiotic stress response and nutrient use efficiency.Science China (Life Sciences), 2020, 63(5): 635-674. |
| 7 | Xie Y P, Chen P X, Yan Y, et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytologist, 2018, 218(1): 201-218. |
| 8 | Samira R, Li B H, Kliebenstein D, et al. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Molecular Biology, 2018, 97(4/5): 297-309. |
| 9 | Govind G, Harshavardhan T G, Patricia J K, et al. Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut. Molecular Genetics and Genomics, 2009, 281(6): 607. |
| 10 | Jin J P, Zhang H, Kong L, et al. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research, 2014, 42: D1182-D1187. |
| 11 | Heim M A, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution, 2003, 20(5): 735-747. |
| 12 | Feller A, Machemer K, Braun E L, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal: For Cell and Molecular Biology, 2011, 66(1): 94-116. |
| 13 | Zhu L L, Zhou B. Regulation of bHLH protein in plant development and abiotic stress. Molecular Plant Breeding, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210222.1744.012.html. |
| 朱璐璐, 周波. bHLH蛋白在植物发育及非生物胁迫中的调控. 分子植物育种, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210222.1744.012.html. | |
| 14 | Ren Y R, Yang Y Y, Zhao Q, et al. MdCIB1, an apple bHLH transcription factor, plays a positive regulator in response to drought stress. Environmental and Experimental Botany, 2021, 188: 1-11. |
| 15 | Pillitteri L J, Torii K U. Breaking the silence: Three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays, 2010, 29(9): 861-870. |
| 16 | Zhao M, Morohashi K, Hatlestad G, et al. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development, 2008, 135(11): 1991-1999. |
| 17 | Abe H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 2003, 15(1): 63-78. |
| 18 | Liu W W, Tai H H, Li S S, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytologist, 2014, 201(4): 1192-1204. |
| 19 | Li H G, Sun J Q, Xu Y X, et al. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Molecular Biology, 2007, 65(5): 655-665. |
| 20 | Min J H, Ju H W, Yoon D, et al. Arabidopsis basic helix-loop-helix 34 (bHLH34) is involved in glucose signaling through binding to a GAGA Cis-element. Frontiers in Plant Science, 2017, 8: 1-14. |
| 21 | Seo J S, Joo J, Kim M J, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. The Plant Journal, 2011, 65(6): 907-921. |
| 22 | Chen C, Liu F, Zhang K X, et al. MeJA-responsive bHLH transcription factor LjbHLH7 regulates cyanogenic glucoside biosynthesis in Lotus japonicus. Journal of Experimental Botany, 2022, 73(8): 2650-2665. |
| 23 | Hou Y Y, Li X, Long R C, et al. Effect of overexpression of the alfalfa MsHB7 gene on drought tolerance of Arabidopsis. Acta Prataculturae Sinica, 2021, 30(4): 170-179. |
| 候怡谣, 李霄, 龙瑞才, 等. 过量表达紫花苜蓿MsHB7基因对拟南芥耐旱性的影响. 草业学报, 2021, 30(4): 170-179. | |
| 24 | Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell, 1989, 56(5): 777-783. |
| 25 | Atchley W R, Fitch W M. A natural classification of the basic helix-loop-helix class of transcription factors. The Proceedings of the National Academy of Sciences, 1997, 94(10): 5172-5176. |
| 26 | Toledo-Ortiz G, Huq E, Quail P H. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell, 2003, 15(8): 1749-1770. |
| 27 | Hao Y Q, Zong X M, Ren P, et al. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. International Journal of Molecular Sciences, 2021, 22(13): 1-20. |
| 28 | Min J H, Park C R, Jang Y H, et al. A basic helix-loop-helix 104 (bHLH104) protein functions as a transcriptional repressor for glucose and abscisic acid signaling in Arabidopsis. Plant Physiology and Biochemistry, 2019, 136: 34-42. |
| 29 | Liu Y J, Ji X Y, Nie X G, et al. AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytologist, 2015, 207(3): 692-709. |
| 30 | Jiang Y, Yang B, Deyholos M K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Molecular Genetics and Genomics, 2009, 282(5): 503-516. |
| 31 | Zhao Q, Fan Z H, Qiu L, et al. MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Frontiers in Plant Science, 2020, 11: 1-16. |
| 32 | Li Z, Liu C, Zhang Y, et al. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. Journal of Experimental Botany, 2019, 70(19): 5471-5486. |
| 33 | Zhang H, Wang P H. Determination of relative water, content in plant leaves in vivo. Plant Physiology Communications, 1991, 27(3): 217-219. |
| 34 | Lanceras J C. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiology, 2004, 135(1): 384-399. |
| 35 | Sudhakar C, Lakshmi A, Giridarakumar S. Changes in the antioxidant enzyme efficacy in two high yielding genotypes of mulberry (Morus alba L.) under NaCl salinity. Plant Science, 2001, 161(3): 613-619. |
| 36 | Pan L. Multi-Omics analysis reveal molecular mechanisms of drought resistance in annual ryegrass (Lolium multiflorum L.). Chengdu: Sichuan Agricultural University, 2018. |
| 潘玲. 多组学联合分析揭示多花黑麦草抗旱响应机制研究. 成都: 四川农业大学, 2018. | |
| 37 | Frugoli J. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiology, 1996, 112(1): 327-336. |
| 38 | Velinov V, Vaseva I, Zehirov G, et al. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Frontiers in Plant Science, 2020, DOI: 10.3389/fpls.2020.00815. |
| 39 | Zou J J, Li X D, Ratnasekera D, et al. Arabidopsis calcium-dependent protein kinase8 and catalase3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. The Plant Cell, 2015, 27(5): 1445-1460. |
| 40 | Wang R S, Pandey S, Li S, et al. Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics, 2011, 12(1): 1-24. |
| [1] | 曾令霜, 李培英, 孙宗玖, 孙晓梵. 两类新疆狗牙根抗旱基因型抗氧化酶保护系统及其基因表达差异分析[J]. 草业学报, 2022, 31(7): 122-132. |
| [2] | 金祎婷, 刘文辉, 刘凯强, 梁国玲, 贾志锋. 全生育期干旱胁迫对‘青燕1号’燕麦叶绿素荧光参数的影响[J]. 草业学报, 2022, 31(6): 112-126. |
| [3] | 苏世平, 李毅, 刘小娥, 种培芳, 单立山, 后有丽. 外源脯氨酸对缓解红砂干旱胁迫的机理研究[J]. 草业学报, 2022, 31(6): 127-138. |
| [4] | 孙晓梵, 张一龙, 李培英, 孙宗玖. 不同施氮量对干旱下狗牙根抗氧化酶活性及渗透调节物质含量的影响[J]. 草业学报, 2022, 31(6): 69-78. |
| [5] | 高莉娟, 张正社, 文裕, 宗西方, 闫启, 卢丽燕, 易显凤, 张吉宇. 象草全基因组bHLH转录因子家族鉴定及表达分析[J]. 草业学报, 2022, 31(3): 47-59. |
| [6] | 王志恒, 魏玉清, 赵延蓉, 王悦娟. 基于转录组学比较研究甜高粱幼苗响应干旱和盐胁迫的生理特征[J]. 草业学报, 2022, 31(3): 71-84. |
| [7] | 高鹏飞, 张静, 范卫芳, 高冰, 郝宏娟, 吴建慧. 干旱胁迫对光叉委陵菜根系特征、结构和生理特性的影响[J]. 草业学报, 2022, 31(2): 203-212. |
| [8] | 吴雨涵, 刘文辉, 刘凯强, 张永超. 干旱胁迫对燕麦幼苗叶片光合特性及活性氧清除系统的影响[J]. 草业学报, 2022, 31(10): 75-86. |
| [9] | 魏娜, 李艳鹏, 马艺桐, 刘文献. 全基因组水平紫花苜蓿TCP基因家族的鉴定及其在干旱胁迫下表达模式分析[J]. 草业学报, 2022, 31(1): 118-130. |
| [10] | 赵颖, 辛夏青, 魏小红. 一氧化氮对干旱胁迫下紫花苜蓿氮代谢的影响[J]. 草业学报, 2021, 30(9): 86-96. |
| [11] | 臧真凤, 白婕, 刘丛, 昝看卓, 龙明秀, 何树斌. 紫花苜蓿形态和生理指标响应干旱胁迫的品种特异性[J]. 草业学报, 2021, 30(6): 73-81. |
| [12] | 罗巧玉, 王彦龙, 陈志, 马永贵, 任启梅, 马玉寿. 水分逆境对发草脯氨酸及其代谢途径的影响[J]. 草业学报, 2021, 30(5): 75-83. |
| [13] | 候怡谣, 李霄, 龙瑞才, 杨青川, 康俊梅, 郭长虹. 过量表达紫花苜蓿MsHB7基因对拟南芥耐旱性的影响[J]. 草业学报, 2021, 30(4): 170-179. |
| [14] | 刘凯强, 刘文辉, 贾志锋, 梁国玲, 马祥. 干旱胁迫对‘青燕1号’燕麦产量及干物质积累与分配的影响[J]. 草业学报, 2021, 30(3): 177-188. |
| [15] | 李冬, 申洪涛, 王艳芳, 王悦华, 王丽君, 赵世民, 刘领. 外源褪黑素对干旱胁迫下烟草幼苗光合碳同化和内源激素的影响[J]. 草业学报, 2021, 30(1): 130-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||