

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (9): 168-182.DOI: 10.11686/cyxb2021347
• 研究论文 • 上一篇
侯丽媛1( ), 贾举庆2, 姜晓东2, 王育川1, 赵菁1, 陈禺怀3, 黄胜雄4(
), 贾举庆2, 姜晓东2, 王育川1, 赵菁1, 陈禺怀3, 黄胜雄4( ), 吴慎杰1(
), 吴慎杰1( ), 董艳辉1(
), 董艳辉1( )
)
收稿日期:2021-09-14
修回日期:2021-12-15
出版日期:2022-09-20
发布日期:2022-08-12
通讯作者:
黄胜雄,吴慎杰,董艳辉
作者简介:Corresponding author. E-mail: dongfang5513@163.com基金资助:
Li-yuan HOU1( ), Ju-qing JIA2, Xiao-dong JIANG2, Yu-chuan WANG1, Jing ZHAO1, Yu-huai CHEN3, Sheng-xiong HUANG4(
), Ju-qing JIA2, Xiao-dong JIANG2, Yu-chuan WANG1, Jing ZHAO1, Yu-huai CHEN3, Sheng-xiong HUANG4( ), Shen-jie WU1(
), Shen-jie WU1( ), Yan-hui DONG1(
), Yan-hui DONG1( )
)
Received:2021-09-14
Revised:2021-12-15
Online:2022-09-20
Published:2022-08-12
Contact:
Sheng-xiong HUANG,Shen-jie WU,Yan-hui DONG
摘要:
WRKY转录因子参与调控植物生长发育和多种胁迫应答,是一类非常重要的植物转录因子。为解析藜麦WRKY基因的进化特征及挖掘响应胁迫的WRKY基因,本研究利用系统的生物信息学方法在全基因组水平对WRKY基因进行了鉴定,并对其染色体定位、分组、系统进化、共线性分析以及多胁迫条件下的表达模式进行了分析。藜麦基因组中鉴定得到90个WRKY基因;划分为3组:Ⅰ组(18个)、Ⅱ组(46个)和Ⅲ组(12个),其中Ⅱ组成员进一步被划分为5个亚组:Ⅱ-a(9个),Ⅱ-b(4个),Ⅱ-c(13个),Ⅱ-d(10个)和Ⅱ-e(10个)。另外,14个WRKY成员因为WRKYGQK短肽的缺失,以及锌指结构变异较大而未划分到任何分组。本研究分组与藜麦WRKY基因进化树中家族成员的聚类结果完全一致,进一步支持了成员分组的可靠性。此外,不同分组的WRKY成员的蛋白序列呈现出小组特异的氨基酸保守域组成。藜麦和祖先二倍体苍白茎藜、瑞典藜的同源基因组模块分析表明,藜麦WRKY基因数目的增加主要来自全基因组倍增。在干旱、高温、盐、低磷胁迫和花生褪绿扇形斑病毒(GCFSV)侵染下,大量WRKY基因的表达水平被显著性诱导或抑制,说明这些WRKY基因很可能参与了调控藜麦的逆境应答反应。研究结果可为藜麦的抗逆研究提供优良的候选WRKY基因,为藜麦的抗逆研究提供参考依据。
侯丽媛, 贾举庆, 姜晓东, 王育川, 赵菁, 陈禺怀, 黄胜雄, 吴慎杰, 董艳辉. 藜麦WRKY基因的进化与多胁迫条件下的转录应答[J]. 草业学报, 2022, 31(9): 168-182.
Li-yuan HOU, Ju-qing JIA, Xiao-dong JIANG, Yu-chuan WANG, Jing ZHAO, Yu-huai CHEN, Sheng-xiong HUANG, Shen-jie WU, Yan-hui DONG. The evolution, characterization and transcriptional responses to multiple stresses of the WRKY genes in Chenopodium quinoa[J]. Acta Prataculturae Sinica, 2022, 31(9): 168-182.
基因名 Gene name | 原始数据库编号 Locus name | 染色体定位 Chromosome location | 蛋白序列长度 Protein sequence length (aa) | WRKY结构域数目 WRKY domain number | 分组 Group |
|---|---|---|---|---|---|
| CqWRKY01 | AUR62021917-RA | Chr01:38208086-38211722 | 554 | 1 | Ⅱ-a |
| CqWRKY02 | AUR62040662-RA | Chr01:41775764-41783826 | 551 | 2 | Ⅰ |
| CqWRKY03 | AUR62024943-RA | Chr01:43875072-43877592 | 169 | 1 | NG |
| CqWRKY04 | AUR62034455-RA | Chr01:57959517-57963772 | 465 | 2 | Ⅰ |
| CqWRKY05 | AUR62009104-RA | Chr01:107241991-107242734 | 163 | 1 | Ⅱ-c |
| CqWRKY06 | AUR62009363-RA | Chr01:110924944-110929629 | 491 | 2 | Ⅰ |
| CqWRKY07 | AUR62023400-RA | Chr01:113698649-113699630 | 84 | 1 | NG |
| CqWRKY08 | AUR62004411-RA | Chr01:118507284-118509071 | 238 | 1 | Ⅱ-e |
| CqWRKY09 | AUR62004415-RA | Chr01:118526620-118532476 | 197 | 1 | NG |
| CqWRKY10 | AUR62004466-RA | Chr01:119081433-119085048 | 373 | 1 | Ⅲ |
| CqWRKY11 | AUR62041828-RA | Chr01:123569655-123571740 | 298 | 1 | Ⅱ-e |
| CqWRKY12 | AUR62027582-RA | Chr01:130910533-130928523 | 354 | 1 | Ⅱ-c |
| CqWRKY13 | AUR62012739-RA | Chr02:1822446-1825621 | 462 | 1 | Ⅱ-a |
| CqWRKY14 | AUR62039475-RA | Chr02:9252429-9253764 | 168 | 1 | NG |
| CqWRKY15 | AUR62017056-RA | Chr02:21804094-21811919 | 580 | 2 | Ⅰ |
| CqWRKY16 | AUR62030160-RA | Chr02:30059138-30065792 | 388 | 1 | Ⅱ-e |
| CqWRKY17 | AUR62030260-RA | Chr02:34031077-34038129 | 252 | 1 | Ⅱ-c |
| CqWRKY18 | AUR62043687-RA | Chr03:29405201-29406855 | 302 | 1 | Ⅱ-d |
| CqWRKY19 | AUR62002702-RA | Chr03:49414886-49421542 | 383 | 1 | Ⅱ-e |
| CqWRKY20 | AUR62012478-RA | Chr03:75647735-75651312 | 299 | 1 | Ⅱ-b |
| CqWRKY21 | AUR62031604-RA | Chr04:1501766-1505059 | 310 | 1 | Ⅱ-b |
| CqWRKY22 | AUR62023044-RA | Chr04:8688937-8691771 | 462 | 2 | Ⅰ |
| CqWRKY23 | AUR62020567-RA | Chr04:14876253-14877758 | 190 | 1 | Ⅱ-c |
| CqWRKY24 | AUR62018123-RA | Chr04:35199821-35203349 | 456 | 2 | Ⅰ |
| CqWRKY25 | AUR62030596-RA | Chr04:48810761-48812831 | 173 | 1 | NG |
| CqWRKY26 | AUR62037715-RA | Chr04:50908753-50909904 | 174 | 1 | Ⅱ-d |
| CqWRKY27 | AUR62015908-RA | Chr05:435350-438098 | 329 | 1 | Ⅲ |
| CqWRKY28 | AUR62015815-RA | Chr05:1471437-1476258 | 430 | 1 | Ⅱ-a |
| CqWRKY29 | AUR62004902-RA | Chr05:66860908-66862973 | 304 | 1 | Ⅱ-d |
| CqWRKY30 | AUR62006655-RA | Chr05:77289532-77293737 | 755 | 2 | Ⅰ |
| CqWRKY31 | AUR62019339-RA | Chr05:79912043-79914657 | 242 | 1 | Ⅲ |
| CqWRKY32 | AUR62028825-RA | Chr06:3898935-3901720 | 454 | 1 | Ⅱ-a |
| CqWRKY33 | AUR62032850-RA | Chr06:6105039-6109784 | 486 | 2 | Ⅰ |
| CqWRKY34 | AUR62003119-RA | Chr06:11861700-11866570 | 314 | 1 | Ⅱ-c |
| CqWRKY35 | AUR62017414-RA | Chr06:42111914-42114982 | 507 | 2 | Ⅰ |
| CqWRKY36 | AUR62006150-RA | Chr07:72108787-72118523 | 562 | 1 | Ⅲ |
| CqWRKY37 | AUR62016230-RA | Chr07:75538327-75540007 | 317 | 1 | Ⅱ-c |
| CqWRKY38 | AUR62018478-RA | Chr07:85862994-85870698 | 1178 | 1 | Ⅲ |
| CqWRKY39 | AUR62006298-RA | Chr07:88930361-88933011 | 454 | 2 | Ⅰ |
| CqWRKY40 | AUR62006427-RA | Chr07:90989279-90992003 | 325 | 1 | Ⅲ |
| CqWRKY41 | AUR62036235-RA | Chr07:112281685-112283961 | 418 | 2 | Ⅰ |
| CqWRKY42 | AUR62011775-RA | Chr08:988047-988819 | 102 | 1 | Ⅱ-c |
| CqWRKY43 | AUR62011795-RA | Chr08:1181350-1182524 | 211 | 1 | Ⅱ-e |
| CqWRKY44 | AUR62011926-RA | Chr08:2678718-2680825 | 294 | 1 | NG |
| CqWRKY45 | AUR62021729-RA | Chr08:8545709-8559517 | 509 | 1 | Ⅱ-a |
| CqWRKY46 | AUR62032212-RA | Chr08:14272865-14273429 | 143 | 1 | NG |
| CqWRKY47 | AUR62029580-RA | Chr08:27816081-27826652 | 222 | 1 | NG |
| CqWRKY48 | AUR62003648-RA | Chr09:4713878-4717289 | 244 | 1 | Ⅱ-c |
| CqWRKY49 | AUR62003924-RA | Chr09:7798854-7801974 | 290 | 1 | Ⅱ-d |
| CqWRKY50 | AUR62024642-RA | Chr09:11682383-11691897 | 424 | 1 | Ⅱ-e |
| CqWRKY51 | AUR62024618-RA | Chr09:11982778-11983536 | 146 | 1 | Ⅱ-c |
| CqWRKY52 | AUR62032241-RA | Chr09:13313236-13317450 | 349 | 1 | NG |
| CqWRKY53 | AUR62023484-RA | Chr10:2033001-2036555 | 371 | 1 | Ⅲ |
| CqWRKY54 | AUR62022617-RA | Chr10:4697680-4699188 | 283 | 1 | Ⅱ-e |
| CqWRKY55 | AUR62022621-RA | Chr10:4717142-4718114 | 212 | 1 | Ⅱ-c |
| CqWRKY56 | AUR62013751-RA | Chr10:13605433-13612166 | 129 | 1 | NG |
| CqWRKY57 | AUR62038804-RA | Chr10:34116524-34117756 | 231 | 1 | Ⅱ-d |
| CqWRKY58 | AUR62020945-RA | Chr11:1752595-1762500 | 1162 | 1 | Ⅲ |
| CqWRKY59 | AUR62039260-RA | Chr11:74163714-74165384 | 388 | 1 | Ⅱ-c |
| CqWRKY60 | AUR62000311-RA | Chr12:3584311-3588565 | 706 | 2 | Ⅰ |
| CqWRKY61 | AUR62001049-RA | Chr12:13518617-13520608 | 333 | 1 | Ⅱ-d |
| CqWRKY62 | AUR62020401-RA | Chr12:55639586-55644905 | 481 | 1 | Ⅱ-a |
| CqWRKY63 | AUR62020314-RA | Chr12:56890585-56891458 | 129 | 1 | Ⅲ |
| CqWRKY64 | AUR62020288-RA | Chr12:57347810-57348994 | 253 | 1 | Ⅱ-d |
| CqWRKY65 | AUR62007485-RA | Chr13:589572-591021 | 286 | 1 | Ⅱ-d |
| CqWRKY66 | AUR62010821-RA | Chr13:7153914-7155613 | 361 | 1 | Ⅲ |
| CqWRKY67 | AUR62010755-RA | Chr13:8078579-8080104 | 297 | 1 | Ⅱ-e |
| CqWRKY68 | AUR62007938-RA | Chr14:2701764-2706207 | 224 | 1 | Ⅱ-c |
| CqWRKY69 | AUR62037832-RA | Chr14:45048099-45055461 | 1058 | 2 | Ⅰ |
| CqWRKY70 | AUR62005736-RA | Chr14:56112914-56116278 | 422 | 1 | Ⅱ-a |
| CqWRKY71 | AUR62005858-RA | Chr14:58268244-58273300 | 487 | 2 | Ⅰ |
| CqWRKY72 | AUR62026996-RA | Chr15:268971-270747 | 296 | 1 | Ⅱ-b |
| CqWRKY73 | AUR62017958-RA | Chr15:21074268-21077689 | 242 | 1 | Ⅱ-c |
| CqWRKY74 | AUR62024046-RA | Chr15:26625857-26629365 | 245 | 1 | NG |
| CqWRKY75 | AUR62015063-RA | Chr15:57542216-57547401 | 500 | 2 | Ⅰ |
| CqWRKY76 | AUR62008370-RA | Chr16:3731574-3745535 | 675 | 1 | Ⅱ-a |
| CqWRKY77 | AUR62038863-RA | Chr16:13862997-13866799 | 589 | 1 | Ⅱ-a |
| CqWRKY78 | AUR62038851-RA | Chr16:14451516-14451878 | 120 | 1 | NG |
| CqWRKY79 | AUR62019754-RA | Chr16:70647711-70649134 | 296 | 1 | Ⅱ-e |
| CqWRKY80 | AUR62019820-RA | Chr16:71712382-71714094 | 361 | 1 | Ⅲ |
| CqWRKY81 | AUR62039941-RA | Chr16:78777309-78778496 | 276 | 1 | Ⅱ-d |
| CqWRKY82 | AUR62026343-RA | Chr17:57960771-57963505 | 446 | 2 | Ⅰ |
| CqWRKY83 | AUR62029778-RA | Chr17:60158005-60160227 | 323 | 1 | Ⅲ |
| CqWRKY84 | AUR62030836-RA | Chr17:83307838-83309577 | 289 | 1 | Ⅱ-b |
| CqWRKY85 | AUR62040434-RA | Chr18:4550853-4554429 | 230 | 1 | NG |
| CqWRKY86 | AUR62009710-RA | Chr18:27460868-27463405 | 492 | 2 | Ⅰ |
| CqWRKY87 | AUR62021101-RA | Chr00:34656729-34658003 | 356 | 1 | Ⅱ-e |
| CqWRKY88 | AUR62038238-RA | Chr00:134168426-134171808 | 338 | 1 | Ⅱ-d |
| CqWRKY89 | AUR62017520-RA | Chr00:135537911-135543127 | 500 | 2 | Ⅰ |
| CqWRKY90 | AUR62044535-RA | Chr00:163451415-163456457 | 138 | 1 | NG |
表1 藜麦WRKY家族成员
Table 1 The WRKY family members in C. quinoa
基因名 Gene name | 原始数据库编号 Locus name | 染色体定位 Chromosome location | 蛋白序列长度 Protein sequence length (aa) | WRKY结构域数目 WRKY domain number | 分组 Group |
|---|---|---|---|---|---|
| CqWRKY01 | AUR62021917-RA | Chr01:38208086-38211722 | 554 | 1 | Ⅱ-a |
| CqWRKY02 | AUR62040662-RA | Chr01:41775764-41783826 | 551 | 2 | Ⅰ |
| CqWRKY03 | AUR62024943-RA | Chr01:43875072-43877592 | 169 | 1 | NG |
| CqWRKY04 | AUR62034455-RA | Chr01:57959517-57963772 | 465 | 2 | Ⅰ |
| CqWRKY05 | AUR62009104-RA | Chr01:107241991-107242734 | 163 | 1 | Ⅱ-c |
| CqWRKY06 | AUR62009363-RA | Chr01:110924944-110929629 | 491 | 2 | Ⅰ |
| CqWRKY07 | AUR62023400-RA | Chr01:113698649-113699630 | 84 | 1 | NG |
| CqWRKY08 | AUR62004411-RA | Chr01:118507284-118509071 | 238 | 1 | Ⅱ-e |
| CqWRKY09 | AUR62004415-RA | Chr01:118526620-118532476 | 197 | 1 | NG |
| CqWRKY10 | AUR62004466-RA | Chr01:119081433-119085048 | 373 | 1 | Ⅲ |
| CqWRKY11 | AUR62041828-RA | Chr01:123569655-123571740 | 298 | 1 | Ⅱ-e |
| CqWRKY12 | AUR62027582-RA | Chr01:130910533-130928523 | 354 | 1 | Ⅱ-c |
| CqWRKY13 | AUR62012739-RA | Chr02:1822446-1825621 | 462 | 1 | Ⅱ-a |
| CqWRKY14 | AUR62039475-RA | Chr02:9252429-9253764 | 168 | 1 | NG |
| CqWRKY15 | AUR62017056-RA | Chr02:21804094-21811919 | 580 | 2 | Ⅰ |
| CqWRKY16 | AUR62030160-RA | Chr02:30059138-30065792 | 388 | 1 | Ⅱ-e |
| CqWRKY17 | AUR62030260-RA | Chr02:34031077-34038129 | 252 | 1 | Ⅱ-c |
| CqWRKY18 | AUR62043687-RA | Chr03:29405201-29406855 | 302 | 1 | Ⅱ-d |
| CqWRKY19 | AUR62002702-RA | Chr03:49414886-49421542 | 383 | 1 | Ⅱ-e |
| CqWRKY20 | AUR62012478-RA | Chr03:75647735-75651312 | 299 | 1 | Ⅱ-b |
| CqWRKY21 | AUR62031604-RA | Chr04:1501766-1505059 | 310 | 1 | Ⅱ-b |
| CqWRKY22 | AUR62023044-RA | Chr04:8688937-8691771 | 462 | 2 | Ⅰ |
| CqWRKY23 | AUR62020567-RA | Chr04:14876253-14877758 | 190 | 1 | Ⅱ-c |
| CqWRKY24 | AUR62018123-RA | Chr04:35199821-35203349 | 456 | 2 | Ⅰ |
| CqWRKY25 | AUR62030596-RA | Chr04:48810761-48812831 | 173 | 1 | NG |
| CqWRKY26 | AUR62037715-RA | Chr04:50908753-50909904 | 174 | 1 | Ⅱ-d |
| CqWRKY27 | AUR62015908-RA | Chr05:435350-438098 | 329 | 1 | Ⅲ |
| CqWRKY28 | AUR62015815-RA | Chr05:1471437-1476258 | 430 | 1 | Ⅱ-a |
| CqWRKY29 | AUR62004902-RA | Chr05:66860908-66862973 | 304 | 1 | Ⅱ-d |
| CqWRKY30 | AUR62006655-RA | Chr05:77289532-77293737 | 755 | 2 | Ⅰ |
| CqWRKY31 | AUR62019339-RA | Chr05:79912043-79914657 | 242 | 1 | Ⅲ |
| CqWRKY32 | AUR62028825-RA | Chr06:3898935-3901720 | 454 | 1 | Ⅱ-a |
| CqWRKY33 | AUR62032850-RA | Chr06:6105039-6109784 | 486 | 2 | Ⅰ |
| CqWRKY34 | AUR62003119-RA | Chr06:11861700-11866570 | 314 | 1 | Ⅱ-c |
| CqWRKY35 | AUR62017414-RA | Chr06:42111914-42114982 | 507 | 2 | Ⅰ |
| CqWRKY36 | AUR62006150-RA | Chr07:72108787-72118523 | 562 | 1 | Ⅲ |
| CqWRKY37 | AUR62016230-RA | Chr07:75538327-75540007 | 317 | 1 | Ⅱ-c |
| CqWRKY38 | AUR62018478-RA | Chr07:85862994-85870698 | 1178 | 1 | Ⅲ |
| CqWRKY39 | AUR62006298-RA | Chr07:88930361-88933011 | 454 | 2 | Ⅰ |
| CqWRKY40 | AUR62006427-RA | Chr07:90989279-90992003 | 325 | 1 | Ⅲ |
| CqWRKY41 | AUR62036235-RA | Chr07:112281685-112283961 | 418 | 2 | Ⅰ |
| CqWRKY42 | AUR62011775-RA | Chr08:988047-988819 | 102 | 1 | Ⅱ-c |
| CqWRKY43 | AUR62011795-RA | Chr08:1181350-1182524 | 211 | 1 | Ⅱ-e |
| CqWRKY44 | AUR62011926-RA | Chr08:2678718-2680825 | 294 | 1 | NG |
| CqWRKY45 | AUR62021729-RA | Chr08:8545709-8559517 | 509 | 1 | Ⅱ-a |
| CqWRKY46 | AUR62032212-RA | Chr08:14272865-14273429 | 143 | 1 | NG |
| CqWRKY47 | AUR62029580-RA | Chr08:27816081-27826652 | 222 | 1 | NG |
| CqWRKY48 | AUR62003648-RA | Chr09:4713878-4717289 | 244 | 1 | Ⅱ-c |
| CqWRKY49 | AUR62003924-RA | Chr09:7798854-7801974 | 290 | 1 | Ⅱ-d |
| CqWRKY50 | AUR62024642-RA | Chr09:11682383-11691897 | 424 | 1 | Ⅱ-e |
| CqWRKY51 | AUR62024618-RA | Chr09:11982778-11983536 | 146 | 1 | Ⅱ-c |
| CqWRKY52 | AUR62032241-RA | Chr09:13313236-13317450 | 349 | 1 | NG |
| CqWRKY53 | AUR62023484-RA | Chr10:2033001-2036555 | 371 | 1 | Ⅲ |
| CqWRKY54 | AUR62022617-RA | Chr10:4697680-4699188 | 283 | 1 | Ⅱ-e |
| CqWRKY55 | AUR62022621-RA | Chr10:4717142-4718114 | 212 | 1 | Ⅱ-c |
| CqWRKY56 | AUR62013751-RA | Chr10:13605433-13612166 | 129 | 1 | NG |
| CqWRKY57 | AUR62038804-RA | Chr10:34116524-34117756 | 231 | 1 | Ⅱ-d |
| CqWRKY58 | AUR62020945-RA | Chr11:1752595-1762500 | 1162 | 1 | Ⅲ |
| CqWRKY59 | AUR62039260-RA | Chr11:74163714-74165384 | 388 | 1 | Ⅱ-c |
| CqWRKY60 | AUR62000311-RA | Chr12:3584311-3588565 | 706 | 2 | Ⅰ |
| CqWRKY61 | AUR62001049-RA | Chr12:13518617-13520608 | 333 | 1 | Ⅱ-d |
| CqWRKY62 | AUR62020401-RA | Chr12:55639586-55644905 | 481 | 1 | Ⅱ-a |
| CqWRKY63 | AUR62020314-RA | Chr12:56890585-56891458 | 129 | 1 | Ⅲ |
| CqWRKY64 | AUR62020288-RA | Chr12:57347810-57348994 | 253 | 1 | Ⅱ-d |
| CqWRKY65 | AUR62007485-RA | Chr13:589572-591021 | 286 | 1 | Ⅱ-d |
| CqWRKY66 | AUR62010821-RA | Chr13:7153914-7155613 | 361 | 1 | Ⅲ |
| CqWRKY67 | AUR62010755-RA | Chr13:8078579-8080104 | 297 | 1 | Ⅱ-e |
| CqWRKY68 | AUR62007938-RA | Chr14:2701764-2706207 | 224 | 1 | Ⅱ-c |
| CqWRKY69 | AUR62037832-RA | Chr14:45048099-45055461 | 1058 | 2 | Ⅰ |
| CqWRKY70 | AUR62005736-RA | Chr14:56112914-56116278 | 422 | 1 | Ⅱ-a |
| CqWRKY71 | AUR62005858-RA | Chr14:58268244-58273300 | 487 | 2 | Ⅰ |
| CqWRKY72 | AUR62026996-RA | Chr15:268971-270747 | 296 | 1 | Ⅱ-b |
| CqWRKY73 | AUR62017958-RA | Chr15:21074268-21077689 | 242 | 1 | Ⅱ-c |
| CqWRKY74 | AUR62024046-RA | Chr15:26625857-26629365 | 245 | 1 | NG |
| CqWRKY75 | AUR62015063-RA | Chr15:57542216-57547401 | 500 | 2 | Ⅰ |
| CqWRKY76 | AUR62008370-RA | Chr16:3731574-3745535 | 675 | 1 | Ⅱ-a |
| CqWRKY77 | AUR62038863-RA | Chr16:13862997-13866799 | 589 | 1 | Ⅱ-a |
| CqWRKY78 | AUR62038851-RA | Chr16:14451516-14451878 | 120 | 1 | NG |
| CqWRKY79 | AUR62019754-RA | Chr16:70647711-70649134 | 296 | 1 | Ⅱ-e |
| CqWRKY80 | AUR62019820-RA | Chr16:71712382-71714094 | 361 | 1 | Ⅲ |
| CqWRKY81 | AUR62039941-RA | Chr16:78777309-78778496 | 276 | 1 | Ⅱ-d |
| CqWRKY82 | AUR62026343-RA | Chr17:57960771-57963505 | 446 | 2 | Ⅰ |
| CqWRKY83 | AUR62029778-RA | Chr17:60158005-60160227 | 323 | 1 | Ⅲ |
| CqWRKY84 | AUR62030836-RA | Chr17:83307838-83309577 | 289 | 1 | Ⅱ-b |
| CqWRKY85 | AUR62040434-RA | Chr18:4550853-4554429 | 230 | 1 | NG |
| CqWRKY86 | AUR62009710-RA | Chr18:27460868-27463405 | 492 | 2 | Ⅰ |
| CqWRKY87 | AUR62021101-RA | Chr00:34656729-34658003 | 356 | 1 | Ⅱ-e |
| CqWRKY88 | AUR62038238-RA | Chr00:134168426-134171808 | 338 | 1 | Ⅱ-d |
| CqWRKY89 | AUR62017520-RA | Chr00:135537911-135543127 | 500 | 2 | Ⅰ |
| CqWRKY90 | AUR62044535-RA | Chr00:163451415-163456457 | 138 | 1 | NG |

图1 藜麦WRKY基因的染色体定位图中A、B后缀标记表示染色体分别来源于苍白茎藜和瑞典藜。The A and B suffix indicate the chromosome originated from C. pallidicaule and C. suecicum, respectively.
Fig.1 The chromosomal location of C. quinoa WRKY genes

图2 藜麦Ⅱ组WRKY成员的WRKY结构域蛋白序列比对46个Ⅱ组WRKY成员的WRKY结构域序列比对结果全部列出。WRKY结构域中保守的氨基酸残基用红色字体标记,变异的WRKYGQK短肽用蓝色字体标记,锌指结构中的半胱氨酸和组氨酸残基用黑色背景标记。The alignment of WRKY domain peptide sequences in the WRKYs of group Ⅱ are all shown. The conserved amino acids in WRKY domains are labeled in red, the WRKYGQK variants are labeled in blue and the residues of cysteine and histidine in zinc-finger are labeled with black background, respectively.
Fig.2 The alignment of WRKY domain peptide sequences in group Ⅱ C. quinoa WRKYs
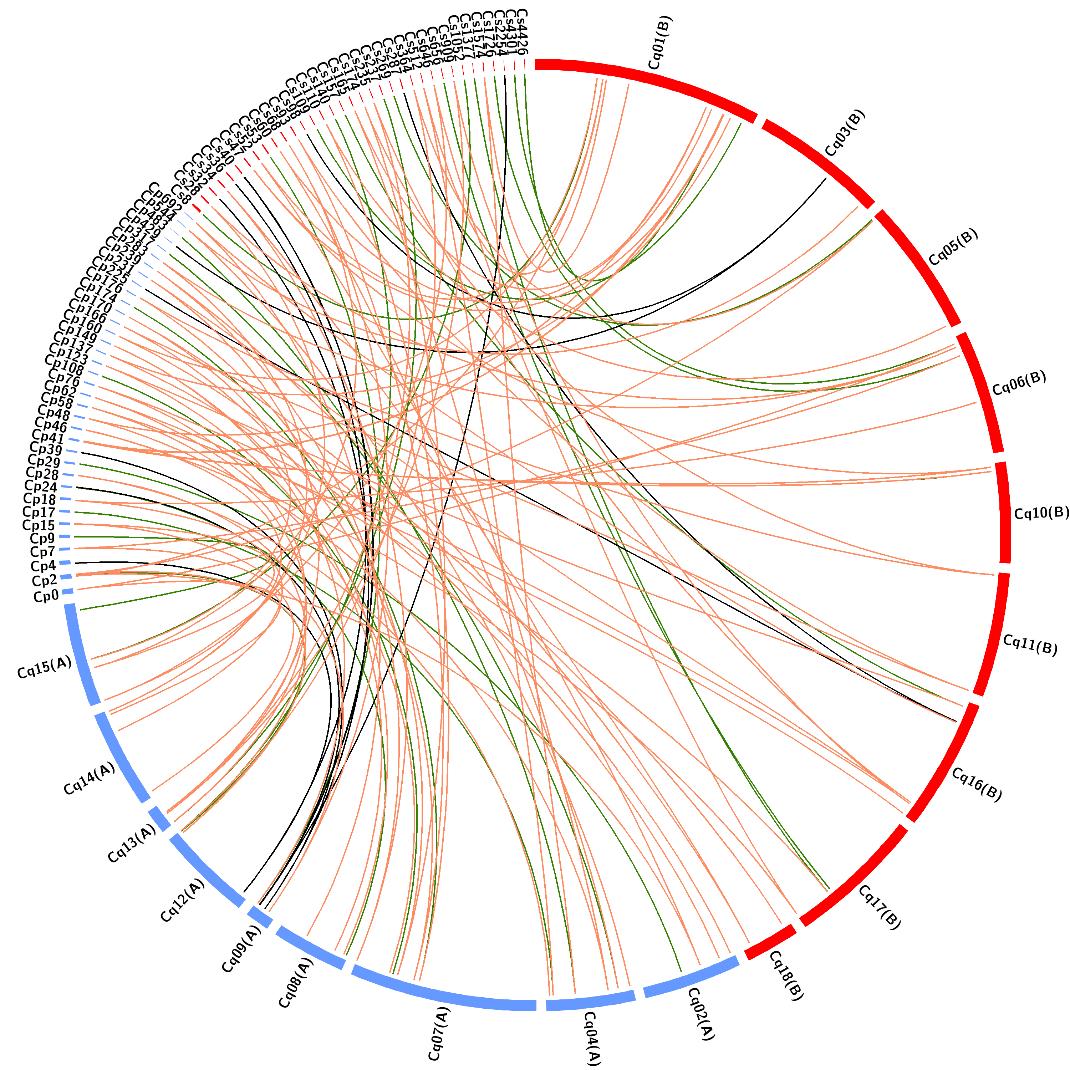
图5 苍白茎藜、瑞典藜和藜麦之间的WRKY基因的保留与丢失苍白茎藜、瑞典藜和藜麦之间所有的包含WRKY基因对的同源基因组模块对。绿色、橙色、黑色的连接线分别表示1 Vs 1、1 Vs 2和2 Vs 1同源关系。All the syntenic genomic region pairs including WRKY gene pairs between C. pallidicaule, C. suecicum and C. quinoa. The green, orange and black links represent the 1 Vs 1, 1 Vs 2, and 2 Vs 1 relationships, respectively.
Fig.5 Reservation and loss of the WRKY genes between C. pallidicaule, C. suecicum and C. quinoa

图6 苍白茎藜、瑞典藜和藜麦之间的WRKY基因的保留与丢失苍白茎藜、瑞典藜和藜麦之间包含同源WRKY基因对的严格意义的1 Vs 2(A)、位置发生改变的1 Vs 2(B)同源基因组模块对。苍白茎藜、瑞典藜和藜麦之间的同源基因组模块对分别用蓝色和红色线条连接。The syntenic genomic region pairs including WRKY gene pairs between C. pallidicaule, C. suecicum, and C. quinoa, including strict 1 Vs 2 (A) and 1 Vs 2 with position arrangement (B) homologous relationships. The links between C. pallidicaule, C. suecicum, and C. quinoa were labeled by blue and red lines, respectively.
Fig.6 Reservation and loss of the WRKY genes between C. pallidicaule, C. suecicum and C. quinoa
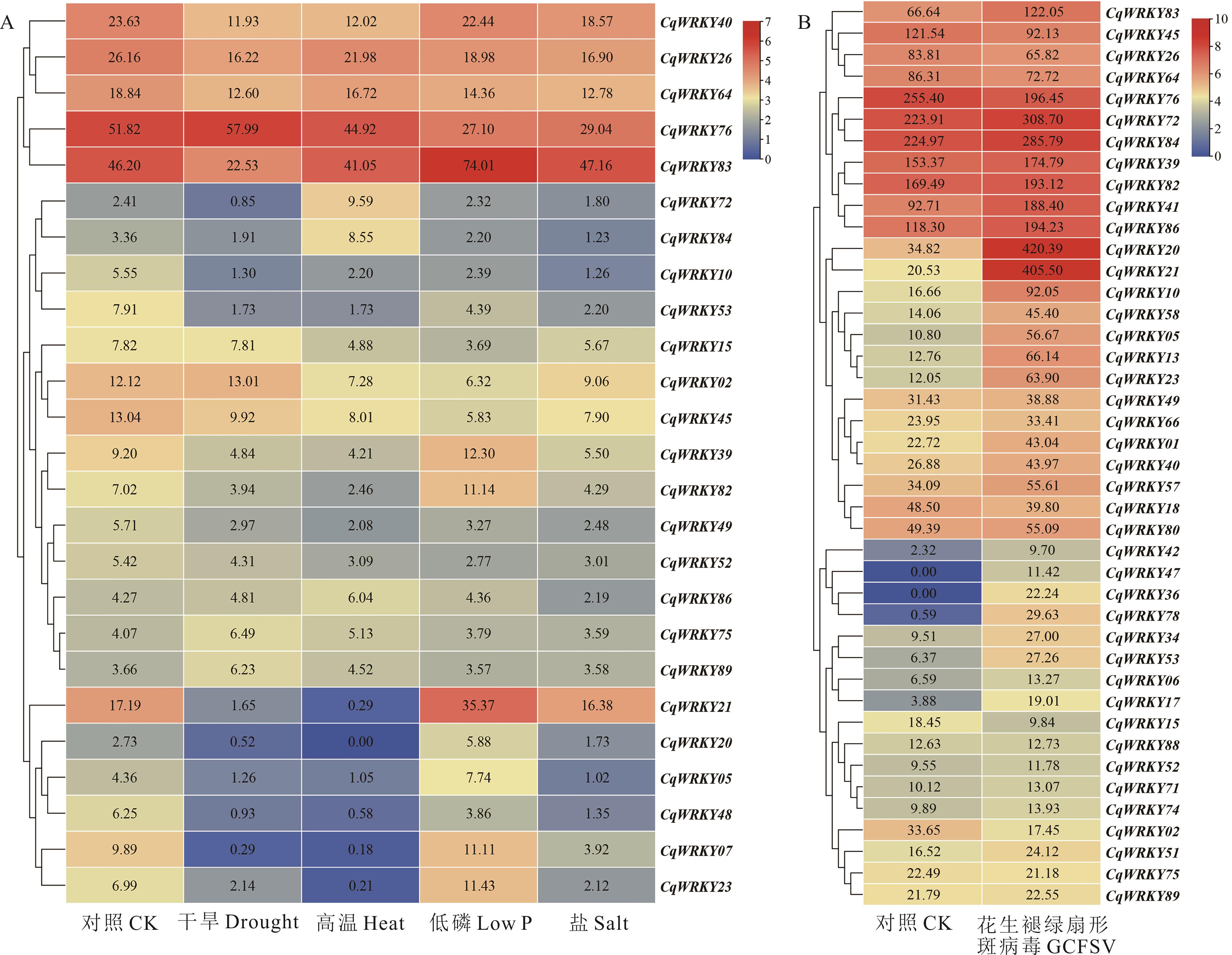
图7 藜麦WRKY基因在干旱、高温、低磷和盐胁迫和GCFSV侵染下的基因表达谱图中基因表达值为TPM值原始数值。图A、B中的WRKY基因在至少一个条件下的表达值分别≥5或≥10。The original TPM data are represented in both figures. The TPM data under at least one condition in figure A and B should be greater than or equal to 5 and 10, respectively.
Fig.7 The CqWRKY genes’ expression profiles under the stresses of drought, heat, low P and salt and GCFSV infection

图8 以葡萄为参照的藜麦WRKY基因的倍增黑色、红色、绿色、蓝色的连接线分别表示1 Vs 1、1 Vs 2、2 Vs 1、多Vs多关系的含有WRKY同源基因的同源基因组模块对。The black, red, green and blue links represent the syntenic genomic regions pairs including homologous WRKYgenes, which indicates 1 Vs 1, 1 Vs 2, 2 Vs 1, and multiple Vs multiple relationships, respectively.
Fig.8 Conservation of the WRKY genes in C. quinoa based on the comparisons to those in V. vinifera
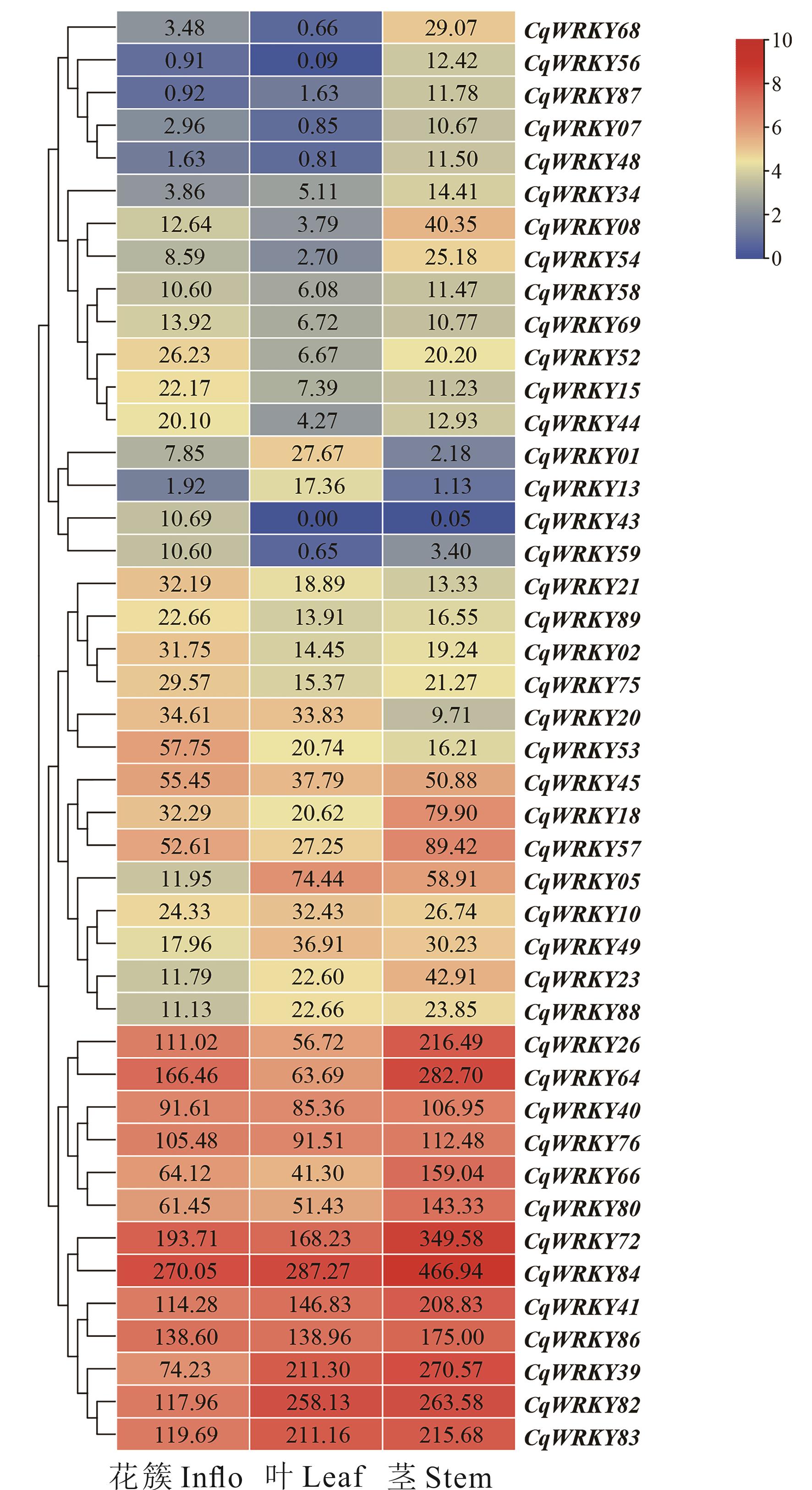
图9 藜麦WRKY基因在花簇、叶和茎中的表达谱图中基因表达值为TPM值原始数值。图中WRKY基因在至少1个组织中的表达值≥10。The original TPM value are shown in the figure. The TPM data in at least one tissue should be greater than or equal to 10, respectively.
Fig.9 The expression profiles of CqWRKY genes in inflorescence, leaf and stem
| 1 | Jarvis D E, Ho Y S, Lightfoot D J, et al. The genome of Chenopodium quinoa. Nature, 2017, 542(7641): 307-312. |
| 2 | Abugoch J L E. Quinoa (Chenopodium quinoa Willd.): Composition, chemistry, nutritional, and functional properties. Advances in Food and Nutrition Research, 2009, 58: 1-31. |
| 3 | Lin M Y, Han P P, Li Y Y, et al. Quinoa secondary metabolites and their biological activities or functions. Molecules, 2019, 24(13): 2512-2559. |
| 4 | Vega-Galvez A, Miranda M, Vergara J, et al. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.), an ancient Andean grain: A review. Journal of the Science of Food and Agriculture, 2010, 90(15): 2541-2547. |
| 5 | Hinojosa L, Gonzalez J A, Barrios-Masias F H, et al. Quinoa abiotic stress responses: A review. Plants, 2018, 7(4): 1-32. |
| 6 | Ulker B, Somssich I E. WRKY transcription factors: From DNA binding towards biological function. Current Opinion in Plant Biology, 2004, 7(5): 491-498. |
| 7 | Rushton P J, Somssich I E, Ringler P, et al. WRKY transcription factors. Trends Plant Science, 2010, 15(5): 247-258. |
| 8 | Eulgem T, Rushton P J, Robatzek S, et al. The WRKY superfamily of plant transcription factors. Trends Plant Science, 2000, 5(5): 199-206. |
| 9 | Rinerson C I, Rabara R C, Tripathi P, et al. The evolution of WRKY transcription factors. BMC Plant Biology, 2015, 15(1): 1-18. |
| 10 | Wani S H, Anand S, Singh B, et al. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Reports, 2021, 40(7): 1071-1085. |
| 11 | Phukan U J, Jeena G S, Shukla R K. WRKY transcription factors: Molecular regulation and stress responses in plants. Frontiers in Plant Science, 2016, 7: 760-774. |
| 12 | Jiang J J, Ma S H, Ye N H, et al. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 2017, 59(2): 86-101. |
| 13 | Wang C T, Ru J N, Liu Y W, et al. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. International Journal of Molecular Science, 2018, 19(10): 3046-3061. |
| 14 | Shi W Y, Du Y T, Ma J, et al. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. International Journal of Molecular Science, 2018, 19(12): 4087-4107. |
| 15 | Gao Y F, Liu J K, Yang F M, et al. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiologia Plantarum, 2020, 168(1): 98-117. |
| 16 | Li X R, Tang Y, Zhou C J, et al. A wheat WRKY transcription factor TaWRKY46 enhances tolerance to osmotic stress in transgenic Arabidopsis plants. International Journal of Molecular Science, 2020, 21(4): 1321-1336. |
| 17 | Zhang D L. Identification and analysis of quinoa MADS-box gene family and study on agrobacterium-mediated root transformation. Yantai: Yantai University, 2021. |
| 张东亮. 藜麦MADS-box基因家族的鉴定与分析及发根农杆菌介导的根转化研究. 烟台: 烟台大学, 2021. | |
| 18 | He L H, Liu S F, Qiao Y, et al. Genome-wide prediction and analysis of quinoa (Chenopodium quinoa) miRNAs and their corresponding target genes. Journal of Shanxi Agricultural University (Natural Science Edition), 2021, 41(1): 1-10. |
| 贺立恒, 刘世芳, 乔宇, 等. 藜麦miRNA及其调控靶基因的全基因组预测与分析. 山西农业大学学报(自然科学版), 2021, 41(1): 1-10. | |
| 19 | Mistry J, Finn R D, Eddy S R, et al. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Research, 2013, 41(12): e121. |
| 20 | Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021. Nucleic Acids Research, 2021, 49(D1): 412-419. |
| 21 | Blum M, Chang H Y, Chuguransky S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Research, 2021, 49(D1): 344-354. |
| 22 | Katoh K, Misawa K, Kuma K, et al. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 2002, 30(14): 3059-3066. |
| 23 | Nguyen L T, Schmidt H A, Haeseler A V, et al. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 2015, 32(1): 268-274. |
| 24 | Kalyaanamoorthy S, Minh B Q, Wong T K F, et al. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 2017, 14(6): 587-589. |
| 25 | Wang Y P, Tang H B, Debarry J D, et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 2012, 40(7): e49. |
| 26 | Krzywinski M, Schein J, Birol I, et al. Circos: An information aesthetic for comparative genomics. Genome Research, 2009, 19: 1639-1645. |
| 27 | Jaillon O, Aury J M, Noel B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 2007, 449(7161): 463-467. |
| 28 | Wang T J, Huang S Z, Zhang A, et al. JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytologist, 2021, 230(2): 567-584. |
| 29 | Pandey S P, Roccaro M, Schön M, et al. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant Journal, 2010, 64(6): 912-923. |
| 30 | Chen H, Lai Z, Shi J, et al. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology, 2010, 10: 281-296. |
| 31 | Aken O V, Zhang B T, Law S, et al. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiology, 2013, 162(1): 254-271. |
| 32 | Li W, Wang H P, Yu D Q. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Molecular Plant, 2016, 9(11): 1492-1503. |
| 33 | Li W, Tian Z X, Yu D Q. WRKY13 acts in stem development in Arabidopsis thaliana. Plant Science, 2015, 236: 205-213. |
| 34 | Sheng Y B, Yan X X, Huang Y, et al. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell and Environment, 2019, 2(3): 891-903. |
| 35 | Zhang Q, Cai W, Ji T T, et al. WRKY13 enhances cadmium tolerance by promoting D-CYSTEINE DESULFHYDRASE and hydrogen sulfide production. Plant Physiology, 2020, 183(1): 345-357. |
| [1] | 苗阳阳, 张艳蕊, 宋标, 刘旭桐, 张安琪, 吕金泽, 张浩, 张小华, 欧阳佳慧, 李旺, 曲善民. 碱蓬根际和内生细菌菌株对盐碱胁迫下苜蓿生长的影响[J]. 草业学报, 2022, 31(9): 107-117. |
| [2] | 苏世平, 刘小娥, 席杰. 三色堇对NaCl胁迫的生理响应[J]. 草业学报, 2022, 31(8): 90-98. |
| [3] | 田骄阳, 王秋霞, 郑淑文, 刘文献. 全基因组水平蒺藜苜蓿CPP基因家族的鉴定及表达模式分析[J]. 草业学报, 2022, 31(7): 111-121. |
| [4] | 曾令霜, 李培英, 孙宗玖, 孙晓梵. 两类新疆狗牙根抗旱基因型抗氧化酶保护系统及其基因表达差异分析[J]. 草业学报, 2022, 31(7): 122-132. |
| [5] | 谢文辉, 黄莉娟, 赵丽丽, 王雷挺, 赵文武. 钙盐胁迫对3份葛藤种质种子萌发及幼苗生理特性的影响[J]. 草业学报, 2022, 31(7): 220-233. |
| [6] | 金祎婷, 刘文辉, 刘凯强, 梁国玲, 贾志锋. 全生育期干旱胁迫对‘青燕1号’燕麦叶绿素荧光参数的影响[J]. 草业学报, 2022, 31(6): 112-126. |
| [7] | 苏世平, 李毅, 刘小娥, 种培芳, 单立山, 后有丽. 外源脯氨酸对缓解红砂干旱胁迫的机理研究[J]. 草业学报, 2022, 31(6): 127-138. |
| [8] | 杨兴云, 乔丹丹, 张雅洁, 王少青, 任俊才, 李明阳, 屈明好, 尚盼盼, 杨成, 黄琳凯, 曾兵. 鸭茅响应水淹胁迫的miRNA差异表达分析[J]. 草业学报, 2022, 31(6): 150-162. |
| [9] | 孙晓梵, 张一龙, 李培英, 孙宗玖. 不同施氮量对干旱下狗牙根抗氧化酶活性及渗透调节物质含量的影响[J]. 草业学报, 2022, 31(6): 69-78. |
| [10] | 刘亚男, 于人杰, 高燕丽, 康俊梅, 杨青川, 武志海, 王珍. 蒺藜苜蓿膜联蛋白MtANN2基因的表达模式及盐胁迫下的功能分析[J]. 草业学报, 2022, 31(5): 124-134. |
| [11] | 张晴, 邢静, 姚佳明, 殷庭超, 黄心如, 何悦, 张敬, 徐彬. 多年生黑麦草细胞分裂素信号通路B类ARR转录因子LpARR10的耐镉功能分析[J]. 草业学报, 2022, 31(5): 135-143. |
| [12] | 欧成明, 赵美琦, 孙铭, 毛培胜. 抗坏血酸和水杨酸丸衣对NaCl胁迫下紫花苜蓿种子发芽特性的影响[J]. 草业学报, 2022, 31(4): 93-101. |
| [13] | 李春杰, 郎鸣晓, 陈振江, 陈泰祥, 刘静, 金媛媛, 魏学凯. Epichloë内生真菌对禾草种子萌发影响研究进展[J]. 草业学报, 2022, 31(3): 192-206. |
| [14] | 王志恒, 魏玉清, 赵延蓉, 王悦娟. 基于转录组学比较研究甜高粱幼苗响应干旱和盐胁迫的生理特征[J]. 草业学报, 2022, 31(3): 71-84. |
| [15] | 高鹏飞, 张静, 范卫芳, 高冰, 郝宏娟, 吴建慧. 干旱胁迫对光叉委陵菜根系特征、结构和生理特性的影响[J]. 草业学报, 2022, 31(2): 203-212. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||