

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (2): 125-137.DOI: 10.11686/cyxb2023106
• 研究论文 • 上一篇
收稿日期:2023-04-06
修回日期:2023-04-24
出版日期:2024-02-20
发布日期:2023-12-12
通讯作者:
马源,王晓丽
作者简介:wxl.yu@163.com基金资助:
Yuan MA1( ), Xiao-li WANG1(
), Xiao-li WANG1( ), Yu-shou MA1, De-gang ZHANG2
), Yu-shou MA1, De-gang ZHANG2
Received:2023-04-06
Revised:2023-04-24
Online:2024-02-20
Published:2023-12-12
Contact:
Yuan MA,Xiao-li WANG
摘要:
为明确高寒草甸退化过程中优势物种改变对根际土壤真菌群落多样性及其稳定性的影响,以青藏高原东缘4个不同退化程度(未退化、轻度退化、中度退化和重度退化)高寒草甸为研究对象,采用ITS rRNA基因测序技术,结合FUNGuild预测和分子生态网络模型方法,分析了高寒草甸退化程度对根际真菌群落结构、功能群和分子生态网络的影响。结果表明:草地退化程度对根际土壤真菌Alpha多样性无显著影响,但显著改变根际土壤真菌的Beta多样性;退化程度仅对根际真菌优势种群的相对丰度产生影响,对真菌优势种群没有影响,4个不同退化程度高寒草甸根际土壤中真菌优势种群均为担子菌门、被孢霉门和子囊菌门;通过线性判别分析发现了29个生物标志物,其中大部分属于担子菌门和子囊菌门;草地退化过程中根际真菌群落主要由共生营养型向腐生营养型转变;网络分析发现,退化高寒草甸根际真菌群落各可操作分类单元(OTU)主要以负相关关系为主,并且退化程度越高负相关程度越强烈。同时结合网络拓扑参数,草地退化程度的加剧将会导致根际真菌结构呈更为松散和不稳定的态势。综上所述,高寒草甸退化程度对优势物种根际真菌群落组成、结构和功能类型产生了显著影响,并降低了真菌群落的稳定性和复杂性。研究结果为深入理解高寒草甸根际微生态的适应性机制提供了科学理论依据。
马源, 王晓丽, 马玉寿, 张德罡. 高寒草甸退化程度对优势物种根际土壤真菌群落和生态网络的影响[J]. 草业学报, 2024, 33(2): 125-137.
Yuan MA, Xiao-li WANG, Yu-shou MA, De-gang ZHANG. Effects of the degree of alpine meadow degradation on the rhizosphere soil fungal community and the ecological network of dominant species[J]. Acta Prataculturae Sinica, 2024, 33(2): 125-137.
样地 Plot | 海拔 Altitude (m) | 纬度 Latitude | 经度 Longitude | 优势物种 Dominant plant species | 盖度 Coverage (%) | 高度 Height (cm) | 地上生物量 Above-ground biomass (g·m-2) |
|---|---|---|---|---|---|---|---|
未退化草地 ND | 3008.3 | 37°13′05″N | 102°44′11″E | 珠芽蓼P.viviparum、垂穗披碱草 E.dahuricus、线叶嵩草K.capillifolia | 98~100 | 20.87 | 403.31 |
轻度退化草地 LD | 2940.0 | 37°11′58″ N | 102°46′17″E | 线叶嵩草K.capillifolia、矮生嵩草 K.humilus、扁蓿豆M.ruthenicus | 82~85 | 18.62 | 364.18 |
中度退化草地 MD | 2869.8 | 37°11′42″N | 102°47′01″E | 矮生嵩草K.humilus、线叶嵩草 K.capillifolia、扁蓿豆M.ruthenicus | 70~78 | 17.38 | 245.42 |
重度退化草地 SD | 2893.6 | 37°12′05″N | 102°45′59″E | 乳白香青A.lactea、矮生嵩草K.humilus、 垂穗披碱草E.dahuricus | 32~38 | 2.16 | 99.05 |
表1 不同退化程度高寒草甸样地基本信息
Table 1 Basic information of alpine meadow plots with different degrees of degradation
样地 Plot | 海拔 Altitude (m) | 纬度 Latitude | 经度 Longitude | 优势物种 Dominant plant species | 盖度 Coverage (%) | 高度 Height (cm) | 地上生物量 Above-ground biomass (g·m-2) |
|---|---|---|---|---|---|---|---|
未退化草地 ND | 3008.3 | 37°13′05″N | 102°44′11″E | 珠芽蓼P.viviparum、垂穗披碱草 E.dahuricus、线叶嵩草K.capillifolia | 98~100 | 20.87 | 403.31 |
轻度退化草地 LD | 2940.0 | 37°11′58″ N | 102°46′17″E | 线叶嵩草K.capillifolia、矮生嵩草 K.humilus、扁蓿豆M.ruthenicus | 82~85 | 18.62 | 364.18 |
中度退化草地 MD | 2869.8 | 37°11′42″N | 102°47′01″E | 矮生嵩草K.humilus、线叶嵩草 K.capillifolia、扁蓿豆M.ruthenicus | 70~78 | 17.38 | 245.42 |
重度退化草地 SD | 2893.6 | 37°12′05″N | 102°45′59″E | 乳白香青A.lactea、矮生嵩草K.humilus、 垂穗披碱草E.dahuricus | 32~38 | 2.16 | 99.05 |
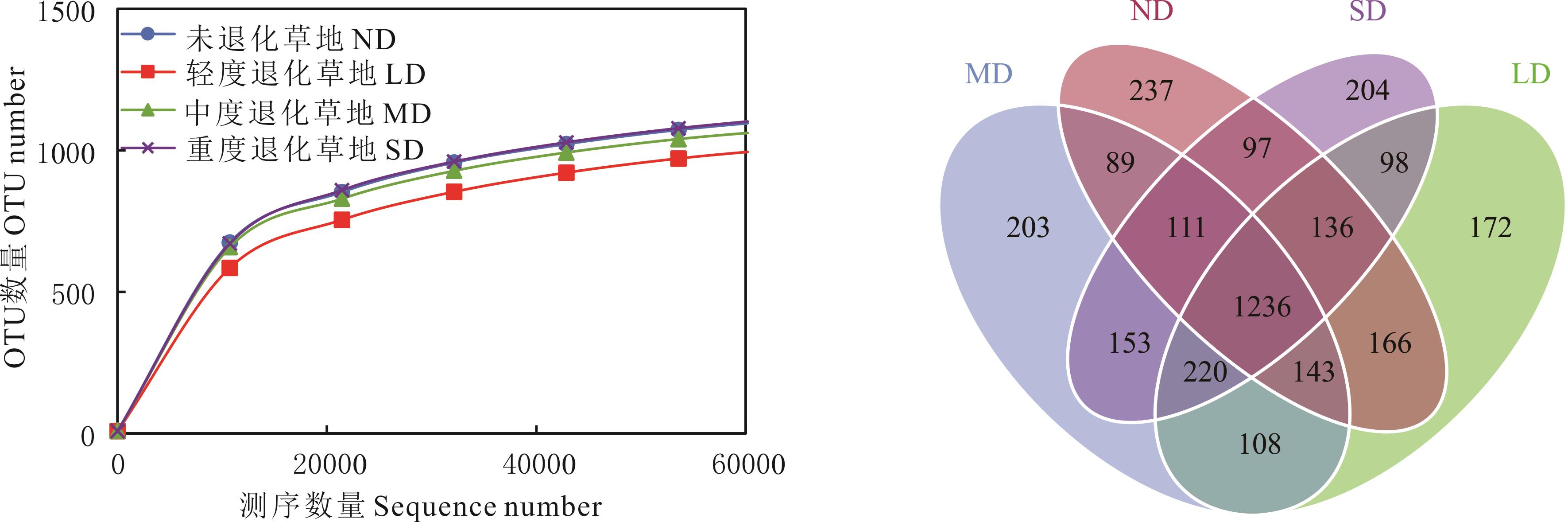
图1 不同退化程度高寒草甸优势物种根际土壤真菌稀释曲线(A)和韦恩图(B)ND: non-degraded grassland, LD: light degraded grassland, MD: moderate degraded grassland, SD: severely degraded grassland. The same below.
Fig.1 Rarefaction curves (A) and Venn diagrams (B) of rhizosphere fungi of dominant species in degraded alpine meadows with different degrees
样品编号 Sample number | Observed_species指数 OTUs | 香农-威纳指数 Shannon-Weiner index | 辛普森指数 Simpson index | Chao1指数 Chao1 index | ACE指数 ACE index | 覆盖度 Coverage (%) |
|---|---|---|---|---|---|---|
| ND | 1121.17±43.70a | 6.77±0.20a | 0.95±0.01a | 1219.75±44.90a | 1230.37±44.90a | 99.72±0.12a |
| LD | 1147.00±87.50a | 6.75±0.63a | 0.92±0.02a | 1284.89±53.80a | 1296.45±52.30a | 99.68±0.11a |
| MD | 1068.50±44.20a | 6.53±0.18a | 0.97±0.01a | 1171.07±44.50a | 1176.99±43.20a | 99.73±0.09a |
| SD | 1063.33±28.10a | 6.51±0.17a | 0.96±0.01a | 1167.33±30.40a | 1175.14±30.90a | 99.74±0.15a |
表 2 不同退化程度高寒草甸优势物种根际土壤真菌多样性指数
Table 2 The rhizosphere soil fungal diversity index of dominant species in degraded alpine meadows with different degrees
样品编号 Sample number | Observed_species指数 OTUs | 香农-威纳指数 Shannon-Weiner index | 辛普森指数 Simpson index | Chao1指数 Chao1 index | ACE指数 ACE index | 覆盖度 Coverage (%) |
|---|---|---|---|---|---|---|
| ND | 1121.17±43.70a | 6.77±0.20a | 0.95±0.01a | 1219.75±44.90a | 1230.37±44.90a | 99.72±0.12a |
| LD | 1147.00±87.50a | 6.75±0.63a | 0.92±0.02a | 1284.89±53.80a | 1296.45±52.30a | 99.68±0.11a |
| MD | 1068.50±44.20a | 6.53±0.18a | 0.97±0.01a | 1171.07±44.50a | 1176.99±43.20a | 99.73±0.09a |
| SD | 1063.33±28.10a | 6.51±0.17a | 0.96±0.01a | 1167.33±30.40a | 1175.14±30.90a | 99.74±0.15a |
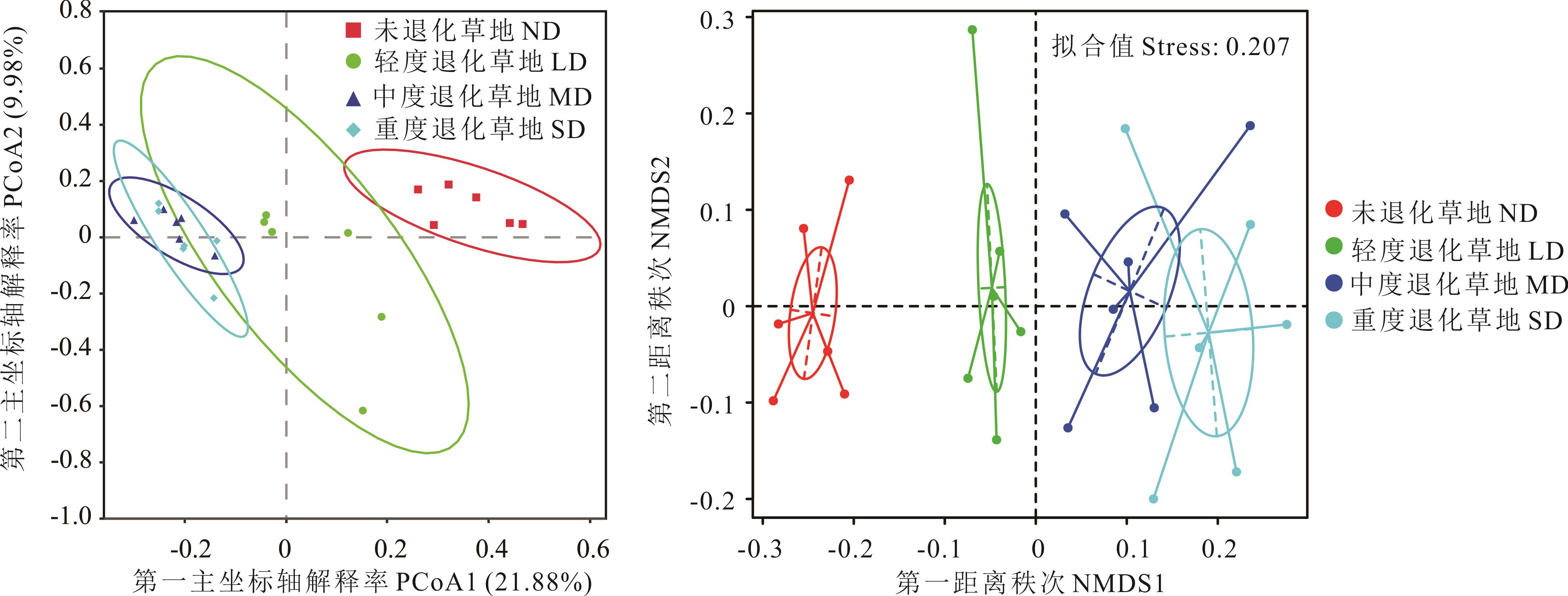
图2 不同退化程度高寒草甸根际土壤真菌OTU水平下PCoA和NMDS分析
Fig.2 Principal co-ordinates analysis(A)and non-metric multi-dimensional scaling analysis (B) based on OTU level of rhizosphere soil fungi in alpine meadows with different degradation degrees

图 3 不同退化程度高寒草甸根际土壤真菌群落门水平相对丰度
Fig.3 Relative abundance of phylum levels of rhizosphere soil fungal communities in alpine meadows with different degrees of degradation
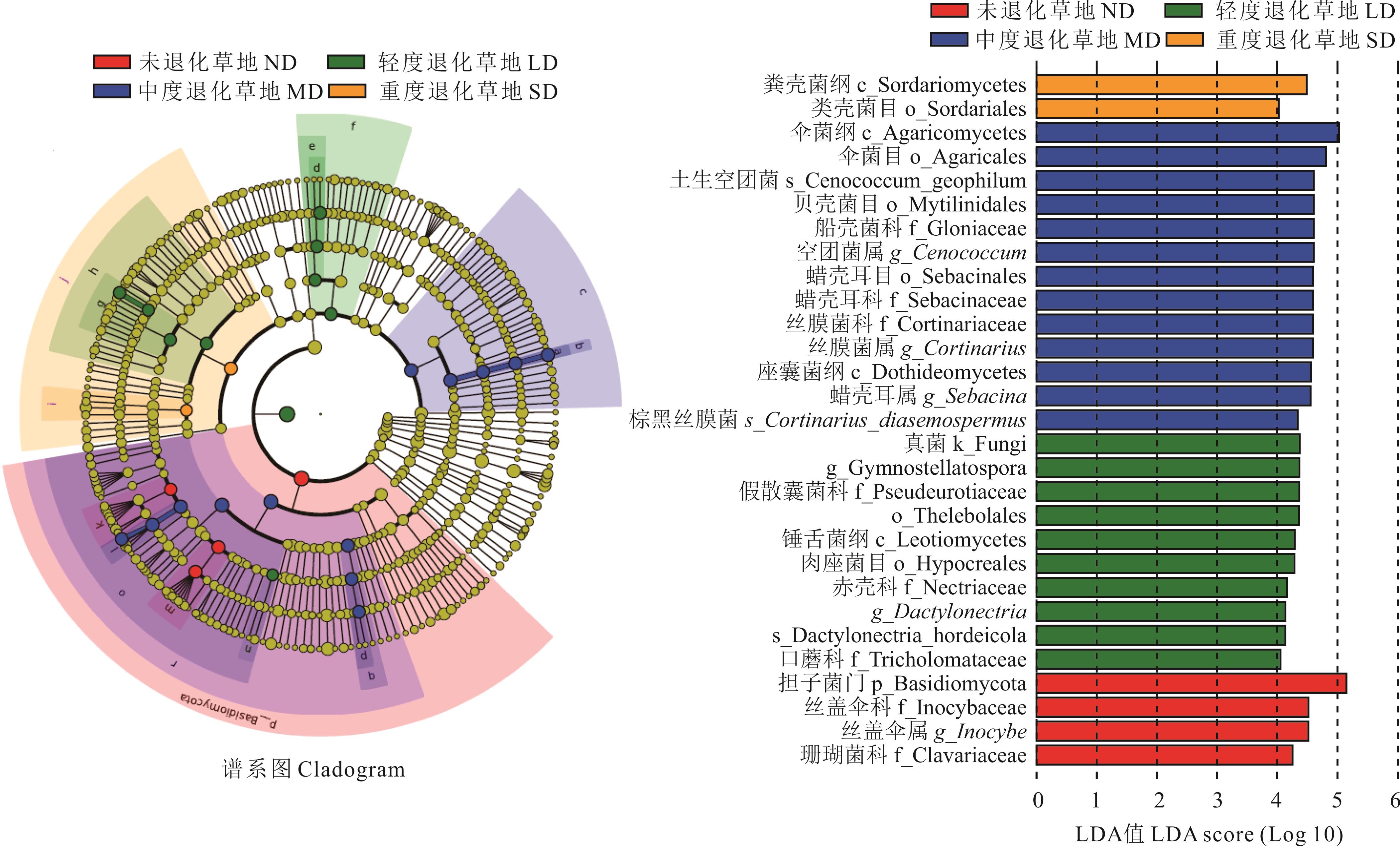
图 4 不同退化程度高寒草甸根际土壤真菌线性判别分析
Fig.4 Linear discriminant analysis effect size of rhizosphere soil fungal community in alpine meadows with different degradation degrees

图 6 不同退化程度高寒草甸根际土壤真菌群落分子生态网络A:ND,未退化草地Non-degraded grassland. B:LD,轻度退化草地Light degraded grassland. C:MD,中度退化草地 Moderate degraded grassland. D: SD,重度退化草地 Severely degraded grassland.下同 The same below. Module:模块性。
Fig.6 Molecular ecological network of rhizosphere soil fungal communities alpine meadows with different degradation degrees
网络 Networks | 项目 Item | 样地 Plot | |||
|---|---|---|---|---|---|
| ND | LD | MD | SD | ||
分子生态网络 Molecular ecological networks | 总节点Total nodes | 839 | 1219 | 971 | 768 |
| 总边Total edges | 6901 | 12929 | 8445 | 5985 | |
| 幂律系数 R2 of power-law | 0.712 | 0.806 | 0.760 | 0.728 | |
| 负相关边比例 Negative edges percentage (%) | 54.31 | 62.19 | 69.37 | 73.92 | |
| 连通性Connectedness | 0.693 | 0.859 | 0.783 | 0.742 | |
| 平均度Average degree | 17.39 | 21.21 | 16.45 | 13.63 | |
| 平均路径长度Average path length | 3.365 | 3.517 | 3.844 | 3.969 | |
| 平均聚集系数Average clustering coefficient | 0.143 | 0.146 | 0.141 | 0.139 | |
| 模块化指数 Modularity | 0.366 | 0.405 | 0.341 | 0.329 | |
| 介数中心性Centralization of betweenness | 0.021 | 0.036 | 0.051 | 0.084 | |
随机网络 Random network | 平均路径长度Average path length | 2.914±0.012 | 2.851±0.010 | 2.929±0.013 | 3.047±0.015 |
| 平均聚集系数Average clustering coefficient | 0.086±0.005 | 0.088±0.003 | 0.084±0.004 | 0.076±0.004 | |
| 模块性Modularity | 0.176±0.002 | 0.151±0.002 | 0.169±0.002 | 0.197±0.003 | |
表 3 不同退化程度高寒草甸根际土壤真菌网络拓扑参数
Table 3 Rhizosphere soil fungal molecular ecological network characteristics of alpine meadow with different degradation degrees
网络 Networks | 项目 Item | 样地 Plot | |||
|---|---|---|---|---|---|
| ND | LD | MD | SD | ||
分子生态网络 Molecular ecological networks | 总节点Total nodes | 839 | 1219 | 971 | 768 |
| 总边Total edges | 6901 | 12929 | 8445 | 5985 | |
| 幂律系数 R2 of power-law | 0.712 | 0.806 | 0.760 | 0.728 | |
| 负相关边比例 Negative edges percentage (%) | 54.31 | 62.19 | 69.37 | 73.92 | |
| 连通性Connectedness | 0.693 | 0.859 | 0.783 | 0.742 | |
| 平均度Average degree | 17.39 | 21.21 | 16.45 | 13.63 | |
| 平均路径长度Average path length | 3.365 | 3.517 | 3.844 | 3.969 | |
| 平均聚集系数Average clustering coefficient | 0.143 | 0.146 | 0.141 | 0.139 | |
| 模块化指数 Modularity | 0.366 | 0.405 | 0.341 | 0.329 | |
| 介数中心性Centralization of betweenness | 0.021 | 0.036 | 0.051 | 0.084 | |
随机网络 Random network | 平均路径长度Average path length | 2.914±0.012 | 2.851±0.010 | 2.929±0.013 | 3.047±0.015 |
| 平均聚集系数Average clustering coefficient | 0.086±0.005 | 0.088±0.003 | 0.084±0.004 | 0.076±0.004 | |
| 模块性Modularity | 0.176±0.002 | 0.151±0.002 | 0.169±0.002 | 0.197±0.003 | |
| 1 | Bardgett R D, Bullock J M, Lavorel S, et al. Combatting global grassland degradation. Nature Reviews Earth and Environment, 2021, 2(10): 720-735. |
| 2 | Zhou H K, Yao B Q, Yu L, et al. Degraded succession and ecological restoration of alpine grassland in the Three-River Source region. Beijing: Science Press, 2016. |
| 周华坤, 姚步青, 于龙, 等. 三江源区高寒草地退化演替与生态恢复. 北京: 科学出版社, 2016. | |
| 3 | Ma Y, Li L Z, Zhang D G, et al. Responses of stoichiometric characteristics of rhizosphere soil to the degradation of alpine meadow. Chinese Journal of Applied Ecology, 2019, 30(9): 3039-3048. |
| 马源, 李林芝, 张德罡, 等. 高寒草甸根际土壤化学计量特征对草地退化的响应. 应用生态学报, 2019, 30(9): 3039-3048. | |
| 4 | Kang L, Chen L, Zhang D, et al. Stochastic processes regulate belowground community assembly in alpine grasslands on the Tibetan Plateau. Environmental Microbiology, 2022, 24(1): 179-194. |
| 5 | Wu G L, Ren G H, Dong Q M, et al. Above- and belowground response along degradation gradient in an alpine grassland of the Qinghai-Tibetan Plateau. CLEAN-Soil, Air, Water, 2014, 42(3): 319-323. |
| 6 | Wagg C, Schlaeppi K, Banerjee S, et al. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nature Communications, 2019, 10(1): 1-10. |
| 7 | Ye F, Wang X, Wang Y, et al. Different pioneer plant species have similar rhizosphere microbial communities. Plant and Soil, 2021, 464(1): 165-181. |
| 8 | Bonanomi G, De Filippis F, Cesarano G, et al. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biology and Biochemistry, 2016, 103: 327-336. |
| 9 | Wu X, Yang J, Ruan H, et al. The diversity and co-occurrence network of soil bacterial and fungal communities and their implications for a new indicator of grassland degradation. Ecological Indicators, 2021, 129: 107989. |
| 10 | He J S, Bu H Y, Hu X W, et al. Close-to-nature restoration of degraded alpine grasslands: Theoretical basis and technical approach. Chinese Science Bulletin, 2020, 65(34): 3898-3908. |
| 贺金生, 卜海燕, 胡小文, 等. 退化高寒草地的近自然恢复: 理论基础与技术途径. 科学通报, 2020, 65(34): 3898-3908. | |
| 11 | Dong L, Li J, Sun J, et al. Soil degradation influences soil bacterial and fungal community diversity in overgrazed alpine meadows of the Qinghai-Tibet Plateau. Scientific Reports, 2021, 11(1): 1-11. |
| 12 | Ren J Z. Scientific research methods of grass industry. Beijing: China Agriculture Press, 1998. |
| 任继周. 草业科学研究方法. 北京: 中国农业出版社, 1998. | |
| 13 | Chaudhary D R, Gautam R K, Yousuf B, et al. Nutrients, microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Applied Soil Ecology, 2015, 91: 16-26. |
| 14 | Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 2012, 41(D1): 590-596. |
| 15 | Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal, 2011, 17(1): 10-12. |
| 16 | Edgar R. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 2013, 10(10): 996-998. |
| 17 | Edgar R C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 2004, 32(5): 1792-1797. |
| 18 | Xiao N, Zhou A, Kempher M L, et al. Disentangling direct from indirect relationships in association networks. Proceedings of the National Academy of Sciences, 2022, 119(2): e2109995119. |
| 19 | Barberán A, Bates S T, Casamayor E O, et al. Using network analysis to explore co-occurrence patterns in soil microbial communities. The ISME Journal, 2012, 6(2): 343-351. |
| 20 | Bastian M, Heymann S, Jacomy M. Gephi: An open source software for exploring and manipulating networks//Proceedings of the international AAAI conference on web and social media. 2009, 3(1): 361-362.https://ojs.aaai.org/index.php/ICWSM/ article/view/13937. |
| 21 | Zhang X, Zhao W, Liu Y, et al. Dominant plant species and soil properties drive differential responses of fungal communities and functions in the soils and roots during secondary forest succession in the subalpine region. Rhizosphere, 2022, 21: 100483. |
| 22 | Bertin C, Yang X, Weston L A. The role of root exudates and allelochemicals in the rhizosphere. Plant and Soil, 2003, 256(1): 67-83. |
| 23 | Dotaniya M L, Meena V D. Rhizosphere effect on nutrient availability in soil and its uptake by plants: A review. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences, 2015, 85(1): 1-12. |
| 24 | Wagg C, Bender S F, Widmer F, et al. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proceedings of the National Academy of Sciences, 2014, 111(14): 5266-5270. |
| 25 | Schmidt P A, Schmitt I, Otte J, et al. Season-long experimental drought alters fungal community composition but not diversity in a grassland soil. Microbial Ecology, 2018, 75(2): 468-478. |
| 26 | Gao X, Dong S, Xu Y, et al. Revegetation significantly increased the bacterial-fungal interactions in different successional stages of alpine grasslands on the Qinghai-Tibetan Plateau. Catena, 2021, 205: 105385. |
| 27 | Yin Y L, Wang Y Q, Li S X, et al. Effects of enclosing on soil microbial community diversity and soil stoichiometric characteristics in a degraded alpine meadow. Chinese Journal of Applied Ecology, 2019, 30(1): 127-136. |
| 尹亚丽, 王玉琴, 李世雄, 等. 围封对退化高寒草甸土壤微生物群落多样性及土壤化学计量特征的影响. 应用生态学报, 2019, 30(1): 127-136. | |
| 28 | Sergio F D A A, Bezerra W M, Dos Santos V M, et al. Fungal diversity in soils across a gradient of preserved Brazilian Cerrado. Journal of Microbiology, 2017, 55(4): 1-7. |
| 29 | Cho N S, Jarosz-Wilkolazka A, Staszczak M, et al. The role of laccase from white rot fungi to stress conditions. Journal Faculty of Agriculture Kyushu University, 2009, 54(1): 81-83. |
| 30 | Vadivelan G, Venkateswaran G. Production and enhancement of omega-3 fatty acid from Mortierella alpina CFR-GV15: Its food and therapeutic application. BioMed Research International, 2014, 2014: 657414. |
| 31 | Ma Y, Zhang D G. Regulation mechanisms of rhizosphere nutrient cycling processes in grassland: A review. Acta Prataculturae Sinica, 2020, 29(11): 172-182. |
| 马源, 张德罡. 草地根际过程对养分循环调控机制研究进展. 草业学报, 2020, 29(11): 172-182. | |
| 32 | Hannula S E, Morrien E, van der Putten W H, et al. Rhizosphere fungi actively assimilating plant-derived carbon in a grassland soil. Fungal Ecology, 2020, 48: 100988. |
| 33 | Floc’h J B, Hamel C, Harker K N, et al. Fungal communities of the canola rhizosphere: Keystone species and substantial between-year variation of the rhizosphere microbiome. Microbial Ecology, 2020, 80(4): 762-777. |
| 34 | Yu Y, Zheng L, Zhou Y, et al. Changes in soil microbial community structure and function following degradation in a temperate grassland. Journal of Plant Ecology, 2021, 14(3): 384-397. |
| 35 | de Vries F T, Griffiths R I, Bailey M, et al. Soil bacterial networks are less stable under drought than fungal networks. Nature Communications, 2018, 9(1): 1-12. |
| 36 | Xu P, Rong X Y, Liu C H, et al. Effects of extreme drought on community and ecological network of soil fungi in a temperate desert. Biodiversity Science, 2022, 30(3): 70-83. |
| 徐鹏, 荣晓莹, 刘朝红, 等. 极端干旱对温带荒漠土壤真菌群落和生态网络的影响. 生物多样性, 2022, 30(3): 70-83. | |
| 37 | Mapelli F, Marasco R, Fusi M, et al. The stage of soil development modulates rhizosphere effect along a high arctic desert chronosequence. The ISME Journal, 2018, 12(5): 1188-1198. |
| 38 | Steele J A, Countway P D, Xia L, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. The ISME Journal, 2011, 5(9): 1414-1425. |
| [1] | 李林芝, 张德罡, 马源, 罗珠珠, 林栋, 海龙, 白兰鸽. 不同退化程度高寒草甸土壤团聚体养分及生态化学计量特征研究[J]. 草业学报, 2023, 32(8): 48-60. |
| [2] | 廖小琴, 王长庭, 刘丹, 唐国, 毛军. 氮磷配施对高寒草甸植物根系特征的影响[J]. 草业学报, 2023, 32(7): 160-174. |
| [3] | 路欣, 祁娟, 师尚礼, 车美美, 李霞, 独双双, 赛宁刚, 贾燕伟. 阔叶类草抑制剂与氮素配施对高寒草甸土壤特性的影响[J]. 草业学报, 2023, 32(7): 38-48. |
| [4] | 刘彩凤, 段媛媛, 王玲玲, 王乙茉, 郭正刚. 高原鼠兔干扰对高寒草甸植物物种多样性与土壤生态化学计量比间关系的影响[J]. 草业学报, 2023, 32(6): 157-166. |
| [5] | 孙玉, 杨永胜, 何琦, 王军邦, 张秀娟, 李慧婷, 徐兴良, 周华坤, 张宇恒. 三江源高寒草甸水源涵养功能及土壤理化性质对退化程度的响应[J]. 草业学报, 2023, 32(6): 16-29. |
| [6] | 周娟娟, 刘云飞, 王敬龙, 魏巍. 短期养分添加对西藏沼泽化高寒草甸地上生物量、植物多样性和功能性状的影响[J]. 草业学报, 2023, 32(11): 17-29. |
| [7] | 韩枫, 张志涛, 张鑫, 王建浩, 王浩. 美国公共牧草地法治管理进程的经验借鉴与若干启示[J]. 草业学报, 2022, 31(9): 220-232. |
| [8] | 游郭虹, 刘丹, 王艳丽, 王长庭. 高寒草甸植物叶片生态化学计量特征对长期氮肥添加的响应[J]. 草业学报, 2022, 31(9): 50-62. |
| [9] | 张玉琢, 杨志贵, 于红妍, 张强, 杨淑霞, 赵婷, 许画画, 孟宝平, 吕燕燕. 基于STARFM的草地地上生物量遥感估测研究——以甘肃省夏河县桑科草原为例[J]. 草业学报, 2022, 31(6): 23-34. |
| [10] | 李洋, 王毅, 韩国栋, 孙建, 汪亚峰. 青藏高原高寒草地土壤微生物量碳氮含量特征及其控制要素[J]. 草业学报, 2022, 31(6): 50-60. |
| [11] | 刘咏梅, 董幸枝, 龙永清, 朱志梅, 王雷, 盖星华, 赵樊, 李京忠. 退化高寒草甸狼毒群落分类特征及其环境影响因子[J]. 草业学报, 2022, 31(4): 1-11. |
| [12] | 李鑫, 魏雪, 王长庭, 任晓, 吴鹏飞. 外源性养分添加对高寒草甸土壤节肢动物群落的影响[J]. 草业学报, 2022, 31(4): 155-164. |
| [13] | 段媛媛, 张静, 王玲玲, 刘彩凤, 王乙茉, 周俗, 郭正刚. 高原鼠兔对高寒草甸植物物种多样性和功能多样性关系的影响[J]. 草业学报, 2022, 31(11): 25-35. |
| [14] | 王永宏, 田黎明, 艾鷖, 陈仕勇, 泽让东科. 短期牦牛放牧对青藏高原高寒草地土壤真菌群落的影响[J]. 草业学报, 2022, 31(10): 41-52. |
| [15] | 唐立涛, 毛睿, 王长庭, 李洁, 胡雷, 字洪标. 氮磷添加对高寒草甸植物群落根系特征的影响[J]. 草业学报, 2021, 30(9): 105-116. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||