

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2022, Vol. 31 ›› Issue (12): 146-157.DOI: 10.11686/cyxb2021478
• 研究论文 • 上一篇
陈倩1,2( ), 徐晓芸1,2, 汪军成1,2, 姚立蓉1,2, 司二静1,2, 杨轲1,2, 韦晓玲1,2, 马小乐1,2, 李葆春1,3, 尚勋武2, 孟亚雄1,2(
), 徐晓芸1,2, 汪军成1,2, 姚立蓉1,2, 司二静1,2, 杨轲1,2, 韦晓玲1,2, 马小乐1,2, 李葆春1,3, 尚勋武2, 孟亚雄1,2( ), 王化俊1,2(
), 王化俊1,2( )
)
收稿日期:2021-12-23
修回日期:2022-02-24
出版日期:2022-12-20
发布日期:2022-10-17
通讯作者:
孟亚雄,王化俊
作者简介:E-mail: huajunwang@sina.com基金资助:
Qian CHEN1,2( ), Xiao-yun XU1,2, Jun-cheng WANG1,2, Li-rong YAO1,2, Er-jing SI1,2, Ke YANG1,2, Xiao-ling WEI1,2, Xiao-le MA1,2, Bao-chun LI1,3, Xun-wu SHANG2, Ya-xiong MENG1,2(
), Xiao-yun XU1,2, Jun-cheng WANG1,2, Li-rong YAO1,2, Er-jing SI1,2, Ke YANG1,2, Xiao-ling WEI1,2, Xiao-le MA1,2, Bao-chun LI1,3, Xun-wu SHANG2, Ya-xiong MENG1,2( ), Hua-jun WANG1,2(
), Hua-jun WANG1,2( )
)
Received:2021-12-23
Revised:2022-02-24
Online:2022-12-20
Published:2022-10-17
Contact:
Ya-xiong MENG,Hua-jun WANG
摘要:
WRKY是一类植物特有的转录因子,尤其在植物生长、发育和逆境过程中起到至关重要的作用。为了鉴定盐生草WRKY基因家族成员、揭示其进化关系及探究HgWRKYs基因在盐胁迫下的表达响应模式,本研究基于盐生草全长转录组数据,利用生物信息学方法对其WRKY转录因子进行鉴定与分析,并通过qRT-PCR方法检测了HgWRKYs家族成员在盐胁迫下的相对表达量。结果表明,在盐生草转录组中共鉴定出31个HgWRKYs,氨基酸数介于220~611 aa,其蛋白N端均具有完整的WRKYGQK保守序列;除HgWRKY20和HgWRKY21同时分布于细胞核和线粒体外,其余HgWRKY基因均位于细胞核;系统进化分析将其分为Ⅰ(8个)、Ⅱ(22个)、Ⅲ(1个)三类,第Ⅱ类进一步分为Ⅱ-a(1个)、Ⅱ-b(2个)、Ⅱ-c(1个)、Ⅱ-d(15个)和Ⅱ-e(3个)5个亚类;Motif 1和Motif 2是HgWRKYs基因家族的特征基序,在进化过程中较为保守。qRT-PCR结果表明,HgWRKYs主要在根系中表达,具有组织特异性,其中HgWRKY19、HgWRKY23和HgWRKY27基因在24 h的表达量最高,将其作为盐胁迫响应候选基因,以期为后续研究盐生草WRKY基因响应盐胁迫的分子机制提供依据。
陈倩, 徐晓芸, 汪军成, 姚立蓉, 司二静, 杨轲, 韦晓玲, 马小乐, 李葆春, 尚勋武, 孟亚雄, 王化俊. 基于全长转录组的盐生草WRKY基因家族的鉴定及其盐胁迫响应模式分析[J]. 草业学报, 2022, 31(12): 146-157.
Qian CHEN, Xiao-yun XU, Jun-cheng WANG, Li-rong YAO, Er-jing SI, Ke YANG, Xiao-ling WEI, Xiao-le MA, Bao-chun LI, Xun-wu SHANG, Ya-xiong MENG, Hua-jun WANG. Identification of a WRKY gene family based on full-length transcriptome sequences and analysis of response patterns under salt stress in Halogeton glomeratus[J]. Acta Prataculturae Sinica, 2022, 31(12): 146-157.
| 基因Gene | 产物长度Product length (bp) | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (5′-3′) |
|---|---|---|---|
| HgWRKY1 | 108 | CGCATCCAAATTGTCCAACGA | TACCAGGCTGAGGCTTAGGA |
| HgWRKY2 | 193 | GCAAGGCACTCAAACCCAAC | CGGTGGTGTCTGACCTTCAT |
| HgWRKY3 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY4 | 136 | CCGCAAGCAAGCAACATGAA | TCCCAACATCACCCACTTCC |
| HgWRKY5 | 108 | CGCATCCAAATTGTCCAACGA | TACCAGGCTGAGGCTTAGGA |
| HgWRKY6 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY7 | 174 | TCCCAGTTGGCTAGGACTGT | CCAGCGGTACCCATCATCAA |
| HgWRKY8 | 120 | CACAACCACGATGTTCCTGC | ACTGAGAGGCCTTAGACCGA |
| HgWRKY9 | 116 | CCATCATCAGGTGGCTCGAA | GCGTTGGTCACGTAGGACTT |
| HgWRKY10 | 162 | TGCCCTCAATGGCTATAAGGC | GCCCAGGGTTTTGTAGCATC |
| HgWRKY11 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY12 | 197 | TCCGATGAAAGAGCCTGGTG | TGGGATGCGGAGTTCCTTTG |
| HgWRKY13 | 102 | TGGATGGAAGTGTGGCGAAT | CGGAGCACCTCTTCCTATGC |
| HgWRKY14 | 193 | TGGATCTTCTGGTCGTTGCC | TCCCCGTACGCTACTACACT |
| HgWRKY15 | 175 | GGGATGGTATCGTCGGTTGG | TCGCGCTTATTGCTGGTACT |
| HgWRKY16 | 113 | GTATCGTCGGTTGGGAGACC | GAACAGTGACAGCGACCAGA |
| HgWRKY17 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY18 | 142 | TCTCTTCGTTGAGCGTGGAT | TTGACACTCCCATCTTCGCC |
| HgWRKY19 | 169 | CCGACAGTGTTCAACCGTCT | TGGCAACGACCAGAAGATCC |
| HgWRKY20 | 195 | CGTTGACTGGTGAGACGGAT | GTCTCCCAACCGACGATACC |
| HgWRKY21 | 119 | GGATGGTATCGTCGGTTGGG | AGAACAGTGACAGCGACCAG |
| HgWRKY22 | 155 | CTTACCCGCACACAAGTCCA | AGACGGTTGAACACTGTCGG |
| HgWRKY23 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY24 | 102 | TGGATGGAAGTGTGGCGAAT | CGGAGCACCTCTTCCTATGC |
| HgWRKY25 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY26 | 191 | GGCGGTGGTGGATCTATGTT | TTACGCTTTGACGAGGAGCA |
| HgWRKY27 | 155 | CTTACCCGCACACAAGTCCA | AGACGGTTGAACACTGTCGG |
| HgWRKY28 | 185 | TACTACCGGTGCACGAACAG | GGCATTGGAGGAGCAAAACG |
| HgWRKY29 | 129 | AGTTACTACCGTTGCACCACT | ACGTGGGAGGACAGTACAAG |
| HgWRKY30 | 163 | ATGGGCAAAAAGCCGTCAAG | AATGTAGCCGGAACTGGGTG |
| HgWRKY31 | 137 | ACAGATGCACCCATCGTCATT | GGGCTGTGAGAAATGGGCTT |
表 1 盐生草WRKY家族基因 qRT-PCR 引物
Table 1 The qRT-PCR primers for WRKY family genes in H. glomeratus
| 基因Gene | 产物长度Product length (bp) | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (5′-3′) |
|---|---|---|---|
| HgWRKY1 | 108 | CGCATCCAAATTGTCCAACGA | TACCAGGCTGAGGCTTAGGA |
| HgWRKY2 | 193 | GCAAGGCACTCAAACCCAAC | CGGTGGTGTCTGACCTTCAT |
| HgWRKY3 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY4 | 136 | CCGCAAGCAAGCAACATGAA | TCCCAACATCACCCACTTCC |
| HgWRKY5 | 108 | CGCATCCAAATTGTCCAACGA | TACCAGGCTGAGGCTTAGGA |
| HgWRKY6 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY7 | 174 | TCCCAGTTGGCTAGGACTGT | CCAGCGGTACCCATCATCAA |
| HgWRKY8 | 120 | CACAACCACGATGTTCCTGC | ACTGAGAGGCCTTAGACCGA |
| HgWRKY9 | 116 | CCATCATCAGGTGGCTCGAA | GCGTTGGTCACGTAGGACTT |
| HgWRKY10 | 162 | TGCCCTCAATGGCTATAAGGC | GCCCAGGGTTTTGTAGCATC |
| HgWRKY11 | 164 | TCGTCGATTTCGGTTGCTGA | ACTACTCTTGGCTCCCTCACT |
| HgWRKY12 | 197 | TCCGATGAAAGAGCCTGGTG | TGGGATGCGGAGTTCCTTTG |
| HgWRKY13 | 102 | TGGATGGAAGTGTGGCGAAT | CGGAGCACCTCTTCCTATGC |
| HgWRKY14 | 193 | TGGATCTTCTGGTCGTTGCC | TCCCCGTACGCTACTACACT |
| HgWRKY15 | 175 | GGGATGGTATCGTCGGTTGG | TCGCGCTTATTGCTGGTACT |
| HgWRKY16 | 113 | GTATCGTCGGTTGGGAGACC | GAACAGTGACAGCGACCAGA |
| HgWRKY17 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY18 | 142 | TCTCTTCGTTGAGCGTGGAT | TTGACACTCCCATCTTCGCC |
| HgWRKY19 | 169 | CCGACAGTGTTCAACCGTCT | TGGCAACGACCAGAAGATCC |
| HgWRKY20 | 195 | CGTTGACTGGTGAGACGGAT | GTCTCCCAACCGACGATACC |
| HgWRKY21 | 119 | GGATGGTATCGTCGGTTGGG | AGAACAGTGACAGCGACCAG |
| HgWRKY22 | 155 | CTTACCCGCACACAAGTCCA | AGACGGTTGAACACTGTCGG |
| HgWRKY23 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY24 | 102 | TGGATGGAAGTGTGGCGAAT | CGGAGCACCTCTTCCTATGC |
| HgWRKY25 | 117 | CATTGCTCCTCGTCAAAGCG | GCCGTATTTGCGCCATGAAT |
| HgWRKY26 | 191 | GGCGGTGGTGGATCTATGTT | TTACGCTTTGACGAGGAGCA |
| HgWRKY27 | 155 | CTTACCCGCACACAAGTCCA | AGACGGTTGAACACTGTCGG |
| HgWRKY28 | 185 | TACTACCGGTGCACGAACAG | GGCATTGGAGGAGCAAAACG |
| HgWRKY29 | 129 | AGTTACTACCGTTGCACCACT | ACGTGGGAGGACAGTACAAG |
| HgWRKY30 | 163 | ATGGGCAAAAAGCCGTCAAG | AATGTAGCCGGAACTGGGTG |
| HgWRKY31 | 137 | ACAGATGCACCCATCGTCATT | GGGCTGTGAGAAATGGGCTT |
基因编号 Genetic code | 基因号 Gene ID | 分子量 Molecular weight (Da) | 氨基酸数 Amino acid quantity (aa) | 长度 Length (bp) | 等电点 Isoelectric point (PI) | 亚细胞定位 Subcellular localization | α螺旋 Alpha-helix (%) | β转角 Beta-turn (%) | 延长链 Extended strand (%) | 无规则卷曲 Random coil (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HgWRKY1 | Hg32527 | 39020.8 | 348 | 1047 | 8.63 | Nucleus | 10.63 | 5.17 | 12.93 | 71.26 |
| HgWRKY2 | Hg33422 | 65872.3 | 611 | 1836 | 6.29 | Nucleus | 9.66 | 3.93 | 13.58 | 72.83 |
| HgWRKY3 | Hg33512 | 64774.9 | 592 | 1779 | 6.48 | Nucleus | 10.81 | 3.38 | 8.78 | 77.03 |
| HgWRKY4 | Hg33871 | 58072.5 | 525 | 1578 | 6.22 | Nucleus | 11.24 | 2.67 | 13.52 | 72.57 |
| HgWRKY5 | Hg35093 | 64054.2 | 578 | 1737 | 6.03 | Nucleus | 11.42 | 2.77 | 10.38 | 75.43 |
| HgWRKY6 | Hg41718 | 48852.7 | 439 | 1320 | 7.61 | Nucleus | 10.48 | 3.87 | 8.43 | 77.22 |
| HgWRKY7 | Hg42748 | 54482.6 | 503 | 1512 | 6.07 | Nucleus | 9.74 | 3.98 | 12.92 | 73.36 |
| HgWRKY8 | Hg47058 | 42592.6 | 380 | 1143 | 8.76 | Nucleus | 11.58 | 2.89 | 13.68 | 71.84 |
| HgWRKY9 | Hg44248 | 40456.7 | 367 | 1104 | 8.43 | Nucleus | 27.25 | 2.18 | 15.80 | 54.77 |
| HgWRKY10 | Hg52691 | 35453.2 | 326 | 981 | 7.14 | Nucleus | 14.72 | 3.07 | 11.66 | 70.55 |
| HgWRKY11 | Hg32535 | 35453.2 | 326 | 981 | 7.14 | Nucleus | 14.72 | 3.07 | 11.66 | 70.55 |
| HgWRKY12 | Hg33409 | 30814.3 | 283 | 852 | 6.80 | Nucleus | 20.85 | 2.47 | 13.43 | 63.25 |
| HgWRKY13 | Hg34478 | 41789.8 | 378 | 1137 | 9.67 | Nucleus | 27.25 | 5.56 | 12.17 | 55.03 |
| HgWRKY14 | Hg34900 | 42129.2 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY15 | Hg40923 | 42243.2 | 387 | 1164 | 9.54 | Nucleus | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY16 | Hg42453 | 42243.2 | 387 | 1164 | 9.54 | Nucleus | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY17 | Hg42552 | 40884.7 | 378 | 1137 | 9.60 | Nucleus | 17.20 | 10.10 | 19.58 | 53.17 |
| HgWRKY18 | Hg43510 | 41710.6 | 377 | 1134 | 9.67 | Nucleus | 28.91 | 4.77 | 10.08 | 56.23 |
| HgWRKY19 | Hg44563 | 42129.4 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY20 | Hg45217 | 42243.2 | 387 | 1164 | 9.54 | Nucleus, Mitochondria | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY21 | Hg46151 | 42243.2 | 387 | 1164 | 9.54 | Nucleus, Mitochondria | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY22 | Hg46356 | 39330.3 | 361 | 1086 | 9.70 | Nucleus | 16.62 | 4.43 | 12.19 | 66.76 |
| HgWRKY23 | Hg46711 | 40884.7 | 378 | 1137 | 9.60 | Nucleus | 17.20 | 10.10 | 19.58 | 53.17 |
| HgWRKY24 | Hg47922 | 41789.8 | 378 | 1137 | 9.67 | Nucleus | 27.25 | 5.56 | 12.17 | 55.03 |
| HgWRKY25 | Hg49441 | 40065.7 | 369 | 1110 | 9.56 | Nucleus | 18.70 | 7.32 | 18.43 | 55.56 |
| HgWRKY26 | Hg50014 | 40610.4 | 375 | 1128 | 9.60 | Nucleus | 17.60 | 8.53 | 19.47 | 54.40 |
| HgWRKY27 | Hg51207 | 42129.4 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY28 | Hg49431 | 24463.3 | 220 | 663 | 9.77 | Nucleus | 9.55 | 4.09 | 13.18 | 73.18 |
| HgWRKY29 | Hg41342 | 45534.9 | 414 | 1245 | 6.19 | Nucleus | 27.78 | 5.07 | 16.91 | 50.24 |
| HgWRKY30 | Hg50291 | 39304.0 | 359 | 1080 | 6.15 | Nucleus | 28.97 | 5.57 | 12.53 | 52.92 |
| HgWRKY31 | Hg45674 | 30599.6 | 272 | 819 | 9.26 | Nucleus | 24.26 | 3.31 | 12.87 | 59.56 |
表2 盐生草WRKY转录因子信息
Table 2 WRKY transcription factors’ information of H. glomeratus
基因编号 Genetic code | 基因号 Gene ID | 分子量 Molecular weight (Da) | 氨基酸数 Amino acid quantity (aa) | 长度 Length (bp) | 等电点 Isoelectric point (PI) | 亚细胞定位 Subcellular localization | α螺旋 Alpha-helix (%) | β转角 Beta-turn (%) | 延长链 Extended strand (%) | 无规则卷曲 Random coil (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HgWRKY1 | Hg32527 | 39020.8 | 348 | 1047 | 8.63 | Nucleus | 10.63 | 5.17 | 12.93 | 71.26 |
| HgWRKY2 | Hg33422 | 65872.3 | 611 | 1836 | 6.29 | Nucleus | 9.66 | 3.93 | 13.58 | 72.83 |
| HgWRKY3 | Hg33512 | 64774.9 | 592 | 1779 | 6.48 | Nucleus | 10.81 | 3.38 | 8.78 | 77.03 |
| HgWRKY4 | Hg33871 | 58072.5 | 525 | 1578 | 6.22 | Nucleus | 11.24 | 2.67 | 13.52 | 72.57 |
| HgWRKY5 | Hg35093 | 64054.2 | 578 | 1737 | 6.03 | Nucleus | 11.42 | 2.77 | 10.38 | 75.43 |
| HgWRKY6 | Hg41718 | 48852.7 | 439 | 1320 | 7.61 | Nucleus | 10.48 | 3.87 | 8.43 | 77.22 |
| HgWRKY7 | Hg42748 | 54482.6 | 503 | 1512 | 6.07 | Nucleus | 9.74 | 3.98 | 12.92 | 73.36 |
| HgWRKY8 | Hg47058 | 42592.6 | 380 | 1143 | 8.76 | Nucleus | 11.58 | 2.89 | 13.68 | 71.84 |
| HgWRKY9 | Hg44248 | 40456.7 | 367 | 1104 | 8.43 | Nucleus | 27.25 | 2.18 | 15.80 | 54.77 |
| HgWRKY10 | Hg52691 | 35453.2 | 326 | 981 | 7.14 | Nucleus | 14.72 | 3.07 | 11.66 | 70.55 |
| HgWRKY11 | Hg32535 | 35453.2 | 326 | 981 | 7.14 | Nucleus | 14.72 | 3.07 | 11.66 | 70.55 |
| HgWRKY12 | Hg33409 | 30814.3 | 283 | 852 | 6.80 | Nucleus | 20.85 | 2.47 | 13.43 | 63.25 |
| HgWRKY13 | Hg34478 | 41789.8 | 378 | 1137 | 9.67 | Nucleus | 27.25 | 5.56 | 12.17 | 55.03 |
| HgWRKY14 | Hg34900 | 42129.2 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY15 | Hg40923 | 42243.2 | 387 | 1164 | 9.54 | Nucleus | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY16 | Hg42453 | 42243.2 | 387 | 1164 | 9.54 | Nucleus | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY17 | Hg42552 | 40884.7 | 378 | 1137 | 9.60 | Nucleus | 17.20 | 10.10 | 19.58 | 53.17 |
| HgWRKY18 | Hg43510 | 41710.6 | 377 | 1134 | 9.67 | Nucleus | 28.91 | 4.77 | 10.08 | 56.23 |
| HgWRKY19 | Hg44563 | 42129.4 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY20 | Hg45217 | 42243.2 | 387 | 1164 | 9.54 | Nucleus, Mitochondria | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY21 | Hg46151 | 42243.2 | 387 | 1164 | 9.54 | Nucleus, Mitochondria | 17.31 | 3.62 | 11.11 | 67.96 |
| HgWRKY22 | Hg46356 | 39330.3 | 361 | 1086 | 9.70 | Nucleus | 16.62 | 4.43 | 12.19 | 66.76 |
| HgWRKY23 | Hg46711 | 40884.7 | 378 | 1137 | 9.60 | Nucleus | 17.20 | 10.10 | 19.58 | 53.17 |
| HgWRKY24 | Hg47922 | 41789.8 | 378 | 1137 | 9.67 | Nucleus | 27.25 | 5.56 | 12.17 | 55.03 |
| HgWRKY25 | Hg49441 | 40065.7 | 369 | 1110 | 9.56 | Nucleus | 18.70 | 7.32 | 18.43 | 55.56 |
| HgWRKY26 | Hg50014 | 40610.4 | 375 | 1128 | 9.60 | Nucleus | 17.60 | 8.53 | 19.47 | 54.40 |
| HgWRKY27 | Hg51207 | 42129.4 | 389 | 1170 | 9.56 | Nucleus | 23.39 | 4.88 | 10.28 | 61.44 |
| HgWRKY28 | Hg49431 | 24463.3 | 220 | 663 | 9.77 | Nucleus | 9.55 | 4.09 | 13.18 | 73.18 |
| HgWRKY29 | Hg41342 | 45534.9 | 414 | 1245 | 6.19 | Nucleus | 27.78 | 5.07 | 16.91 | 50.24 |
| HgWRKY30 | Hg50291 | 39304.0 | 359 | 1080 | 6.15 | Nucleus | 28.97 | 5.57 | 12.53 | 52.92 |
| HgWRKY31 | Hg45674 | 30599.6 | 272 | 819 | 9.26 | Nucleus | 24.26 | 3.31 | 12.87 | 59.56 |

图1 盐生草WRKY蛋白家族序列比对图中字母为氨基酸,颜色无意义(颜色种类使图片便于观察分析),字母大小高低表示序列保守程度,字母越大越高代表保守性越高。The letters in the Figure are amino acids, and the color is meaningless (the color variety makes the picture easy to observe and analyze). The letter size indicates the conservatism of the sequence. The larger the letter, the higher the letter is, the higher the conservatism is.
Fig.1 Sequence alignment of the WRKY protein family in H. glomeratus

图2 盐生草与拟南芥WRKY家族基因进化关系黄色方块标记拟南芥WRKY家族基因,红色三角形标记盐生草WRKY家族基因。Yellow squares mark WRKY family genes of A. thaliana and red triangles mark WRKY family genes of H. glomeratus.
Fig.2 Evolution of WRKY family genes between H. glomeratus and A. thaliana
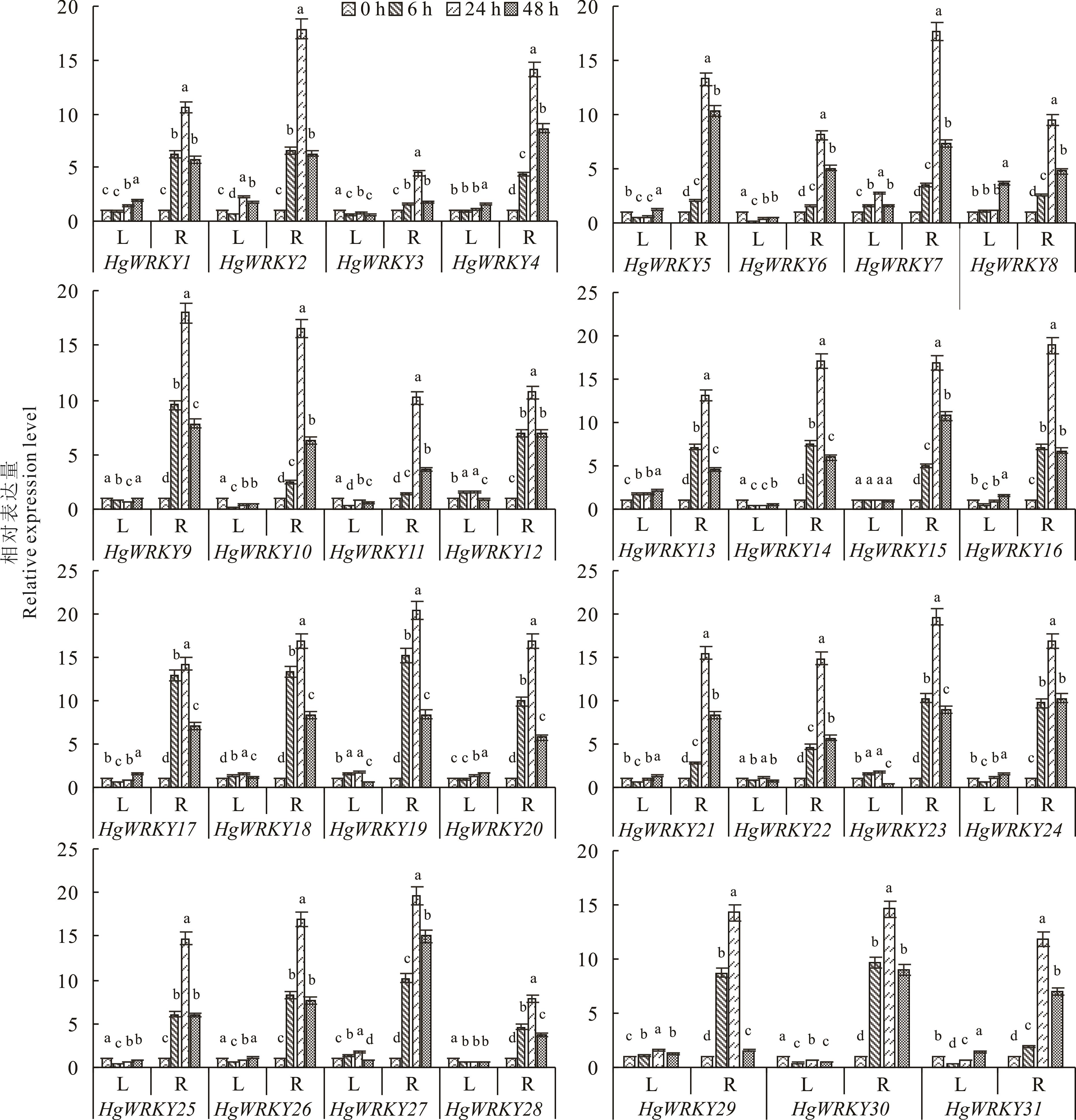
图4 31个HgWRKYs在200 mmol·L-1 NaCl胁迫下不同时间不同组织中的表达量L: 叶片Leaf; R: 根系Root; 图中数据是3次重复的平均值,小写字母表示HgWRKYs在同一组织不同处理时间下差异显著(P<0.05)。The data is the average of three repetitions, the differernt lowercase letters indicate that HgWRKYs have significant differences at different treatment times in the same tissue (P<0.05).
Fig.4 Expression levels of 31 HgWRKYs in different tissues at different times under 200 mmol·L-1 NaCl stress
| 1 | Wicke B, Smeets E, Dornburg V, et al. The global technical and economic potential of bioenergy from salt-affected soils. Energy and Environmental Science, 2011, 4: 2669-2681. |
| 2 | Kaashyap M, Ford R, Bohra A, et al. Improving salt tolerance of chickpea using modern genomics tools and molecular breeding. Current Genomics, 2017, 18(6): 557-567. |
| 3 | Yang J S, Yao R J. Management and efficient agricultural utilization of salt-affected soil in China. Bulletin of Chinese Academy of Sciences, 2015, 30(Z1): 162-170. |
| 杨劲松, 姚荣江. 我国盐碱地的治理与农业高效利用. 中国科学院院刊, 2015, 30(Z1): 162-170. | |
| 4 | Li K, Zhou C J. Research progress in WRKY transcription factors in plants. Plant Physiology Journal, 2014, 50(9): 1329-1335. |
| 李岢, 周春江. 植物WRKY转录因子的研究进展. 植物生理学报, 2014, 50(9): 1329-1335. | |
| 5 | Duan M R, Nan J, Liang Y H, et al. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Research, 2007, 35(4): 1145-1154. |
| 6 | Thomas E, Paul J R, Silke R, et al. The WRKY superfamily of plant transcription factors. Trends in Plant Science, 2000, 5(5): 196-206. |
| 7 | Dou L L, Zhang X H, Pang C Y, et al. Genome-wide analysis of the WRKY gene family in cotton. Molecular Genetics and Genomics, 2014, 289(6): 1103-1121. |
| 8 | Bi C Y, Huang X F, Wang H S, et al. Identification of WRKY transcription factor gene in Ipomoea batatas genome and expression analysis under stresses. Journal of Northwest A&F University (Natural Science Edition), 2021, 49(9): 30-44. |
| 毕楚韵, 黄小芳, 王和寿, 等. 甘薯全基因组WRKY转录因子的基因鉴定与逆境胁迫表达分析. 西北农林科技大学学报(自然科学版), 2021, 49(9): 30-44. | |
| 9 | Jiang T, Lin Y X, Liu X, et al. Genome-wide analysis of WRKY transcription factor genes in alfalfa. Acta Prataculturae Sinica, 2011, 20(3): 211-218. |
| 江腾, 林勇祥, 刘雪, 等. 苜蓿全基因组WRKY转录因子基因的分析. 草业学报, 2011, 20(3): 211-218. | |
| 10 | Zheng J J, Zhang Z L, Tong T, et al. Genome-wide identification of WRKY gene family and expression analysis under abiotic stress in barley. Agronomy, 2021, 11(3): 521. |
| 11 | Johnson C S, Kolevski B, Smyth D R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. The Plant Cell, 2002, 14(6): 1359-1375. |
| 12 | Wang Q J, Chen L G, Yu D Q. Overexpression of AtWRKY71 affects plant’s defense response to Pseudomonas syringae. Plant Diversity and Resources, 2015, 37(5): 577-585. |
| 王其娟, 陈利钢, 余迪求. 过表达AtWRKY71影响植物对病原菌Pseudomonas syringae的抗性. 植物分类与资源学报, 2015, 37(5): 577-585. | |
| 13 | Zhou X, Jiang Y, Yu D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecules and Cells, 2011, 31(4): 303-313. |
| 14 | Sakamoto H, Maruyama K, Sakuma Y, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold and high-salinity stress conditions. Plant Physiology, 2004, 136(1): 2734-2746. |
| 15 | Wang R Y, Wen W W, Zhao E H, et al. Cloning and salt-tolerance analysis of MsWRKY11 in alfalfa. Acta Prataculturae Sinica, 2021, 30(11): 157-169. |
| 王如月, 文武武, 赵恩华, 等. 紫花苜蓿MsWRKY11基因的克隆及其耐盐功能分析. 草业学报, 2021, 30(11): 157-169. | |
| 16 | Benckemalato M, Cabreira C, Wiebkestrohm B, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biology, 2014, 14(1): 1-18. |
| 17 | Yao L R, Wang J C, Li B C, et al. Influences of heavy metals and salt on seed germination and seedling characteristics of halophyte Halogeton glomeratus. Bulletin of Environmental Contamination and Toxicology, 2021, 106(3): 545-556. |
| 18 | Wang J C, Meng Y C, Li B C, et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell & Environment, 2015, 38(4): 655-669. |
| 19 | Wang J C, Yao L R, Li B C, et al. Single-molecule long-read transcriptome dataset of halophyte Halogeton glomeratus. Frontiers in Genetics, 2017, 8: 197. |
| 20 | Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 2013, 30(12): 2725-2729. |
| 21 | Ma Y H, Xu X L, Wang J C, et al. Cloning and expression analysis of Actin gene fragment from halophyte Halogeton glomeratus. Pratacultural Science, 2015, 32(9): 1432-1437. |
| 马艳红, 徐先良, 汪军成, 等. 盐生草Actin基因片段的克隆及表达. 草业科学, 2015, 32(9): 1432-1437. | |
| 22 | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative and the 2-ΔΔCT method. Methods, 2001, 25(4): 402-408. |
| 23 | Chen X, Chen R H, Wang Y F, et al. Genome-wide identification of WRKY transcription factors in Chinese jujube (Ziziphus jujuba Mill.) and their involvement in fruit developing, ripening, and abiotic stress. Genes, 2019, 10(5): 360. |
| 24 | Sun S H, Yu D Q. WRKY transcription factors in regulation of stress response in plant. Biotechnology Bulletin, 2016, 32(10): 66-76. |
| 孙淑豪, 余迪求. WRKY转录因子家族调控植物逆境胁迫响应. 生物技术通报, 2016, 32(10): 66-76. | |
| 25 | Kong W L, Yu K, Dan N Z, et al. Genome-wide identification and expression analysis of WRKY transcription factor under abiotic stress in Beta vulgaris. Scientia Agricultura Sinica, 2017, 50(17): 3259-3273. |
| 孔维龙, 于坤, 但乃震, 等. 甜菜WRKY转录因子全基因组鉴定及其在非生物胁迫下的表达分析. 中国农业科学, 2017, 50(17): 3259-3273. | |
| 26 | Ross C A, Liu Y, Shen Q J. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology, 2007, 49(6): 827-842. |
| 27 | Huang S, Gao Y, Liu J, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics, 2012, 287(6): 495-513. |
| 28 | Li M Y, Xu Z S, Tian C, et al. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Scientific Reports, 2016, 6(1): 1-17. |
| 29 | Zhang Y, Wang L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evolution Biology, 2005, 5(1): 1-11. |
| 30 | Lin S N, Liu J W, Zhang X N, et al. Genome-wide identification and expression analysis of WRKY gene family in Dianthus caryophyllus. Acta Horticulturae Sinica, 2021, 48(9): 1768-1784. |
| 林胜男, 刘杰玮, 张晓妮, 等. 香石竹WRKY家族全基因组鉴定及其表达分析. 园艺学报, 2021, 48(9): 1768-1784. | |
| 31 | Che Y M, Sun Y J, Lu S C, et al. AtWRKY40 functions in drought stress response in Arabidopsis thaliana. Plant Physiology Journal, 2018, 54(3): 456-464. |
| 车永梅, 孙艳君, 卢松冲, 等. AtWRKY40参与拟南芥干旱胁迫响应过程. 植物生理学报, 2018, 54(3): 456-464. | |
| 32 | Chen L G, Zhang L P, Li D B, et al. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the USA, 2013, 110(21): 1963-1971. |
| 33 | Tong C C, Zhang C, Fan T T, et al. Cloning and expression analysis of AtWRKY33 gene promoter from Arabidopsis thaliana. Journal of Anhui Agricultural University, 2020, 47(5): 784-788. |
| 童晨晨, 张乘, 樊婷婷, 等. 拟南芥AtWRKY33基因启动子的克隆及表达分析. 安徽农业大学学报, 2020, 47(5): 784-788. | |
| 34 | Bakshi M, Oelmüller R. WRKY transcription factors: Lack of many trades in plants. Plant Signaling & Behavior, 2014, 9 (2): e27700. |
| 35 | Van Aken O, Zhang B, Law S, et al. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiology, 2013, 162: 254-271. |
| 36 | Hruz T, Laule O, Szabo G, et al. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics, 2008, 2008: 35-39. |
| 37 | Vanderauwera S, Vandenbroucke K, Inzé A, et al. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the USA, 2012, 109(49): 20113-20118. |
| 38 | Muhammad A A, Farrukh A, Muhammad A N, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. Journal of Plant Physiology, 2018, 226: 12-21. |
| [1] | 谢文辉, 黄莉娟, 赵丽丽, 王雷挺, 赵文武. 钙盐胁迫对3份葛藤种质种子萌发及幼苗生理特性的影响[J]. 草业学报, 2022, 31(7): 220-233. |
| [2] | 刘亚男, 于人杰, 高燕丽, 康俊梅, 杨青川, 武志海, 王珍. 蒺藜苜蓿膜联蛋白MtANN2基因的表达模式及盐胁迫下的功能分析[J]. 草业学报, 2022, 31(5): 124-134. |
| [3] | 王志恒, 魏玉清, 赵延蓉, 王悦娟. 基于转录组学比较研究甜高粱幼苗响应干旱和盐胁迫的生理特征[J]. 草业学报, 2022, 31(3): 71-84. |
| [4] | 李娜娜, 刘同歌, 黄志慧, 郑宝江, 张玉红. 草本资源植物菥蓂对盐胁迫下生理生态及次生代谢产物响应[J]. 草业学报, 2022, 31(11): 181-190. |
| [5] | 张鹏, 任茜, 孟思宇, 魏小星, 鲍根生. 内生真菌对盐胁迫下紫花针茅种子萌发和幼苗生长的研究[J]. 草业学报, 2022, 31(10): 110-121. |
| [6] | 吴彤, 刘云苗, 金军, 董伟峰, 才晓溪, 孙明哲, 贾博为, 孙晓丽. 蒺藜苜蓿cation/H+ exchanger基因家族鉴定及表达特征分析[J]. 草业学报, 2022, 31(1): 181-194. |
| [7] | 陆安桥, 张峰举, 许兴, 王学琴, 姚姗. 盐胁迫对湖南稷子苗期生长及生理特性的影响[J]. 草业学报, 2021, 30(5): 84-93. |
| [8] | 汪芳珍, 杨成行, 何子华, 林子茹, 曾浩源, 马清. 盐处理下旱生植物沙芥蛋白激酶相关基因的差异表达分析[J]. 草业学报, 2021, 30(10): 116-124. |
| [9] | 田甜, 王海江, 王金刚, 朱永琪, 史晓艳, 李维弟, 李文瑞玉. 盐胁迫下施加氮素对饲用油菜有机渗透调节物质积累的影响[J]. 草业学报, 2021, 30(10): 125-136. |
| [10] | 王苗苗, 周向睿, 梁国玲, 赵桂琴, 焦润安, 柴继宽, 高雪梅, 李娟宁. 5份燕麦材料苗期耐盐性综合评价[J]. 草业学报, 2020, 29(8): 143-154. |
| [11] | 黄勇, 郭猛, 张红瑞, 周艳, 李贺敏, 高致明, 王盼盼. 盐胁迫对石竹种子萌发和幼苗生长的影响[J]. 草业学报, 2020, 29(12): 105-111. |
| [12] | 王桔红, 史生晶, 陈文, 甘桂媚, 陈赛娜, 李张伟. 枯草芽孢杆菌和3种放线菌对盐胁迫下鬼针草和鳢肠种子萌发及幼苗生长的影响[J]. 草业学报, 2020, 29(12): 112-120. |
| [13] | 何建军, 姚立蓉, 汪军成, 边秀秀, 司二静, 杨轲, 王化俊, 马小乐, 李葆春, 尚勋武, 孟亚雄. 干旱和盐胁迫对盐生植物盐生草种子萌发特性的影响[J]. 草业学报, 2020, 29(11): 129-140. |
| [14] | 李珍, 云岚, 石子英, 王俊, 张晨, 郭宏宇, 盛誉. 盐胁迫对新麦草种子萌发及幼苗期生理特性的影响[J]. 草业学报, 2019, 28(8): 119-129. |
| [15] | 伍国强, 李辉, 雷彩荣, 蔺丽媛, 金娟, 李善家. 添加KCl对高盐胁迫下红豆草生长及生理特性的影响[J]. 草业学报, 2019, 28(6): 45-55. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||