

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2023, Vol. 32 ›› Issue (4): 129-141.DOI: 10.11686/cyxb2022180
• 研究论文 • 上一篇
收稿日期:2022-04-20
修回日期:2022-06-27
出版日期:2023-04-20
发布日期:2023-01-29
通讯作者:
徐彬
作者简介:E-mail: binxu@njau.edu.cn基金资助:
Jia-ming YAO( ), Huan-huan HAO, Jing ZHANG, Bin XU(
), Huan-huan HAO, Jing ZHANG, Bin XU( )
)
Received:2022-04-20
Revised:2022-06-27
Online:2023-04-20
Published:2023-01-29
Contact:
Bin XU
摘要:
tRNA可以将多个sgRNAs连接起来合并成一个多顺反子基因,再与CRISPR/Cas9表达载体结合形成多顺反子tRNA-sgRNA/Cas9(PTG/Cas9)系统来对多靶点进行基因编辑。该系统已经在水稻中验证其可促进sgRNAs的转录和提高多靶点的编辑效率。因此,为了在多年生黑麦草中实现高效的基因编辑,本研究中,构建了2个带有tRNA的CRISPR中间载体,并提供了一种快速、灵活的PTG/Cas9载体构建方法。为了快速验证PTG/Cas9系统是否可以在多年生黑麦草基因组中发挥功能,用聚乙二醇4000(PEG 4000)介导PTG/Cas9质粒转化多年生黑麦草原生质体,后提取原生质体DNA扩增目的序列检测是否有目标基因被编辑的细胞。试验结果显示,PTG/Cas9系统可用于多年生黑麦草基因编辑,且基因编辑效率约为6.7%。试验结果为多年生黑麦草遗传研究和育种提供了重要的基础。
姚佳明, 郝欢欢, 张敬, 徐彬. tRNA-sgRNA/Cas9系统介导多年生黑麦草原生质体的基因编辑[J]. 草业学报, 2023, 32(4): 129-141.
Jia-ming YAO, Huan-huan HAO, Jing ZHANG, Bin XU. The use of the tRNA-sgRNA/Cas9 system for gene editing in perennial ryegrass protoplasts[J]. Acta Prataculturae Sinica, 2023, 32(4): 129-141.
| 引物Primer | 引物序列Primer sequence (5′-3′) |
|---|---|
| S1F | TTGCAGTATGGGCCGGCCCATTACG |
| S1R | ACGTTGTAAAACGACGGCCAGTGCC |
| S2F | CCATGATTACGAATTCGAGCTCGGTACCCGG |
| S2R | CTGCACATCTGATTCCTCCAAGATCCAT |
| F1 | CTCCGTTTTACCTGTGGAATC |
| R1 | |
| F2 | |
| R2 | CGGAGGAAAATTCCATCCAC |
| F3 | GGCTCGTATGTTGTGTGG |
| R3 | |
| F4 | |
| F5 | TTCAGAGGTCTCTACCGTGGAATCGGCAGCAAA |
| R5 | TTCAGAGGTCTCA |
| F6 | GAGGTCTCG |
| R6 | AGCGTGGGTCTCGCTCGTCCATCCACTCCAAGC |
| F7 | GCGAGCCTTATAAGCAG |
| R7 | GCTGAGATAACGAGCCAA |
表1 试验所用引物
Table 1 Primers used in this study
| 引物Primer | 引物序列Primer sequence (5′-3′) |
|---|---|
| S1F | TTGCAGTATGGGCCGGCCCATTACG |
| S1R | ACGTTGTAAAACGACGGCCAGTGCC |
| S2F | CCATGATTACGAATTCGAGCTCGGTACCCGG |
| S2R | CTGCACATCTGATTCCTCCAAGATCCAT |
| F1 | CTCCGTTTTACCTGTGGAATC |
| R1 | |
| F2 | |
| R2 | CGGAGGAAAATTCCATCCAC |
| F3 | GGCTCGTATGTTGTGTGG |
| R3 | |
| F4 | |
| F5 | TTCAGAGGTCTCTACCGTGGAATCGGCAGCAAA |
| R5 | TTCAGAGGTCTCA |
| F6 | GAGGTCTCG |
| R6 | AGCGTGGGTCTCGCTCGTCCATCCACTCCAAGC |
| F7 | GCGAGCCTTATAAGCAG |
| R7 | GCTGAGATAACGAGCCAA |
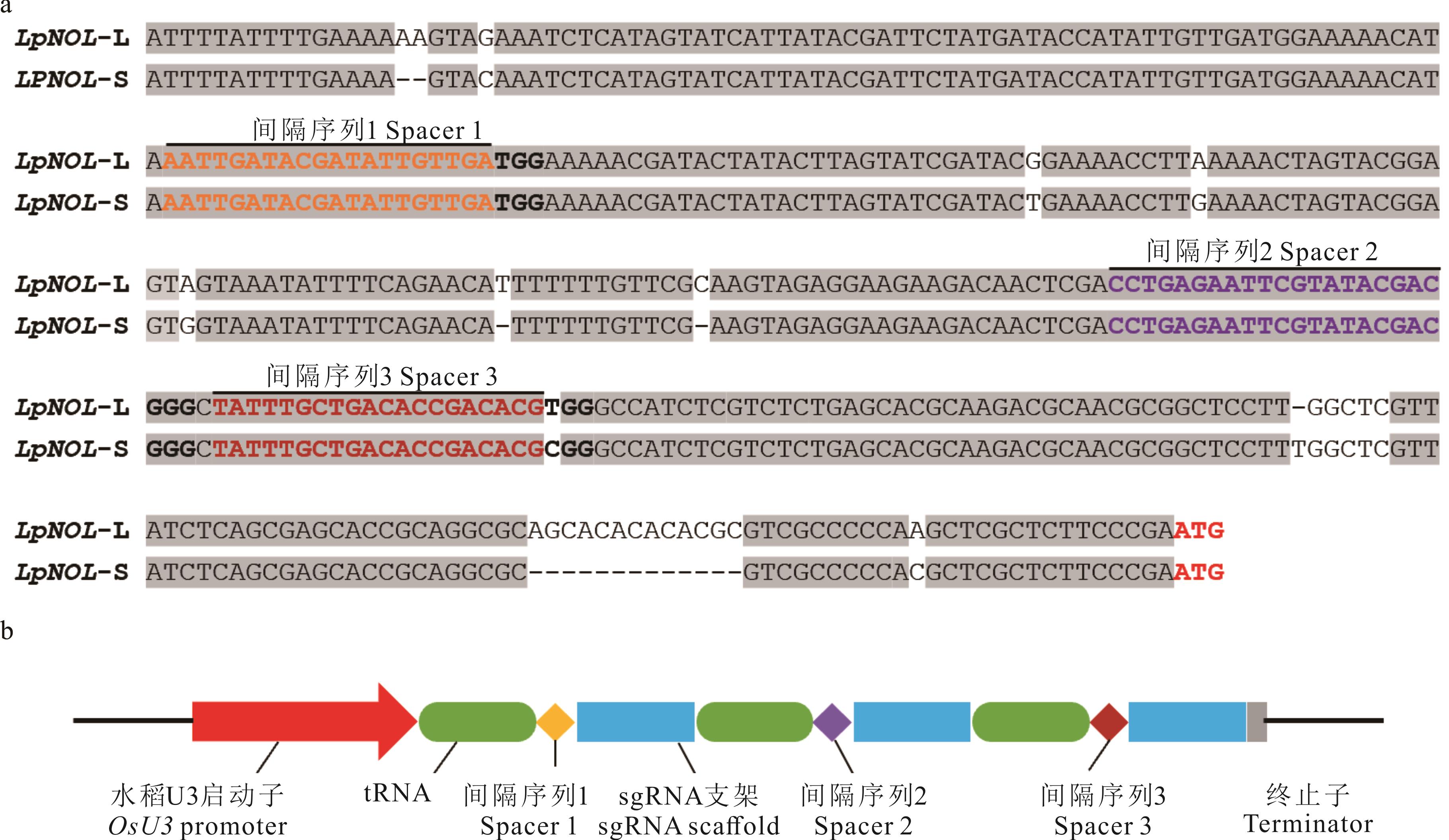
图1 sgRNA在多年生黑麦草基因组中的位置(a)及tRNA-sgRNA多顺反子(PTG)表达盒的结构(b)黄色字体碱基序列代表间隔序列1,紫色代表间隔序列2,红棕色代表间隔序列3。下同。Yellow colored nucleotides represented the spacer 1, purple ones represented the spacer 2, reddish-brown ones represented the spacer 3. The same below.
Fig.1 Location of sgRNAs in the genome of perennial ryegrass and the structure of the polycistronic tRNA-sgRNA (PTG) cassette
| 步骤Steps | 反应组分Reaction components | 组分名称Component name | 体积Volume (μL) |
|---|---|---|---|
PCR组分 Components of the PCR | PCR缓冲液PCR buffer | 2×Phanta Max buffer | 15.0 |
| DRT | dNTP Mix (10 mmol·L-1) | 0.6 | |
| 模板Template | POT/PT (100 ng·L-1) | 1.0 | |
| 上游引物Forward primer | F1/F2/F3/F4 (10 μmol·L-1) | 1.2 | |
| 下游引物Reverse primer | R1/R2/R3/R2 (10 μmol·L-1) | 1.2 | |
| DNA聚合酶DNA polymerase | Phanta Max Super-Fidelity DNA polymerase | 0.6 | |
| 双蒸水Double distilled water | ddH2O | 10.4 | |
| 共计Total | 30.0 | ||
重叠式PCR Overlapping PCR | PCR缓冲液PCR buffer | 2×Phanta Max buffer | 15.0 |
| DRT | dNTP Mix (10 mmol·L-1) | 0.6 | |
| 模板Template | 产物1、产物2/产物3、产物4 Product 1, product 2/product 3, product 4 | 1.0+1.0 | |
| 上游引物Forward primer | F5/F6 (10 μmol·L-1) | 1.2 | |
| 下游引物Reverse primer | R5/R6 (10 μmol·L-1) | 1.2 | |
| DNA聚合酶DNA polymerase | Phanta Max Super-Fidelity DNA polymerase | 0.6 | |
| 双蒸水Double distilled water | ddH2O | 9.4 | |
| 共计Total | 30.0 | ||
金门组装反应 Golden gate assembly | 缓冲液Buffer | 10×CutSmart | 1.0 |
| 10×T4 DNA ligase buffer | 1.0 | ||
| 连接产物Products for connection | 产物5 Product 5 | 2.5 | |
| 产物6 Product 6 | 2.5 | ||
| 表达载体Expression vector | pYLCRISPR Cas9Pubi-H | 2.0 | |
| 限制性内切酶Restriction enzyme | Bsa I | 0.5 | |
| 连接酶Ligase | T4 DNA ligase | 0.5 | |
| 共计Total | 10.0 |
表2 PCR反应体系
Table 2 PCR reaction mix
| 步骤Steps | 反应组分Reaction components | 组分名称Component name | 体积Volume (μL) |
|---|---|---|---|
PCR组分 Components of the PCR | PCR缓冲液PCR buffer | 2×Phanta Max buffer | 15.0 |
| DRT | dNTP Mix (10 mmol·L-1) | 0.6 | |
| 模板Template | POT/PT (100 ng·L-1) | 1.0 | |
| 上游引物Forward primer | F1/F2/F3/F4 (10 μmol·L-1) | 1.2 | |
| 下游引物Reverse primer | R1/R2/R3/R2 (10 μmol·L-1) | 1.2 | |
| DNA聚合酶DNA polymerase | Phanta Max Super-Fidelity DNA polymerase | 0.6 | |
| 双蒸水Double distilled water | ddH2O | 10.4 | |
| 共计Total | 30.0 | ||
重叠式PCR Overlapping PCR | PCR缓冲液PCR buffer | 2×Phanta Max buffer | 15.0 |
| DRT | dNTP Mix (10 mmol·L-1) | 0.6 | |
| 模板Template | 产物1、产物2/产物3、产物4 Product 1, product 2/product 3, product 4 | 1.0+1.0 | |
| 上游引物Forward primer | F5/F6 (10 μmol·L-1) | 1.2 | |
| 下游引物Reverse primer | R5/R6 (10 μmol·L-1) | 1.2 | |
| DNA聚合酶DNA polymerase | Phanta Max Super-Fidelity DNA polymerase | 0.6 | |
| 双蒸水Double distilled water | ddH2O | 9.4 | |
| 共计Total | 30.0 | ||
金门组装反应 Golden gate assembly | 缓冲液Buffer | 10×CutSmart | 1.0 |
| 10×T4 DNA ligase buffer | 1.0 | ||
| 连接产物Products for connection | 产物5 Product 5 | 2.5 | |
| 产物6 Product 6 | 2.5 | ||
| 表达载体Expression vector | pYLCRISPR Cas9Pubi-H | 2.0 | |
| 限制性内切酶Restriction enzyme | Bsa I | 0.5 | |
| 连接酶Ligase | T4 DNA ligase | 0.5 | |
| 共计Total | 10.0 |
| 常规PCR Regular PCR | 重叠式PCR Overlapping PCR | 金门组装Golden gate assembly | ||||||
|---|---|---|---|---|---|---|---|---|
| 步骤Step | 温度Temperature (℃) | 时间Time (s) | 步骤Step | 温度Temperature (℃) | 时间Time (s) | 步骤Step | 温度Temperature (℃) | 时间Time (s) |
| 1 | 95 | 180 | 1 | 95 | 180 | 1 | 37 | 300 |
| 2 | 95 | 15 | 2 | 95 | 15 | 2 | 16 | 600 |
| 3 | 60 | 15 | 3 | 60 | 15 | 3 | - | 7200 |
| 4 | 72 | 30 | 4 | 72 | 30 | |||
| 5 | - | 2100 | 5 | - | 2100 | |||
| 6 | 72 | 300 | 6 | 72 | 3000 | |||
表3 PCR反应条件
Table 3 PCR reaction condition
| 常规PCR Regular PCR | 重叠式PCR Overlapping PCR | 金门组装Golden gate assembly | ||||||
|---|---|---|---|---|---|---|---|---|
| 步骤Step | 温度Temperature (℃) | 时间Time (s) | 步骤Step | 温度Temperature (℃) | 时间Time (s) | 步骤Step | 温度Temperature (℃) | 时间Time (s) |
| 1 | 95 | 180 | 1 | 95 | 180 | 1 | 37 | 300 |
| 2 | 95 | 15 | 2 | 95 | 15 | 2 | 16 | 600 |
| 3 | 60 | 15 | 3 | 60 | 15 | 3 | - | 7200 |
| 4 | 72 | 30 | 4 | 72 | 30 | |||
| 5 | - | 2100 | 5 | - | 2100 | |||
| 6 | 72 | 300 | 6 | 72 | 3000 | |||
| 溶液Solution | 成分Ingredient |
|---|---|
| 酶解液Enzyme solution | 10 mmol·L-1 MES,1.5%纤维素酶R10 Cellulase R10,0.75%离析酶R10 Macerozyme R10,20 mmol·L-1 KCl,10 mmol·L-1 CaCl2,0.1% BSA,0.6 mol·L-1甘露醇Mannitol |
| W5 | 154 mmol·L-1 NaCl,125 mmol·L-1 CaCl2,5 mmol·L-1 KCl,2 mmol·L-1 MES |
| MMg | 0.4 mol·L-1甘露醇Mannitol,15 mmol·L-1 MgCl2,4 mmol·L-1 MES |
| PEG 4000 | 20% PEG 4000,10 mmol·L-1 CaCl2,0.3 mol·L-1甘露醇Mannitol |
表4 多年生黑麦草原生质体提取及转化所用溶液
Table 4 Solutions used to extract and transform perennial ryegrass protoplasts
| 溶液Solution | 成分Ingredient |
|---|---|
| 酶解液Enzyme solution | 10 mmol·L-1 MES,1.5%纤维素酶R10 Cellulase R10,0.75%离析酶R10 Macerozyme R10,20 mmol·L-1 KCl,10 mmol·L-1 CaCl2,0.1% BSA,0.6 mol·L-1甘露醇Mannitol |
| W5 | 154 mmol·L-1 NaCl,125 mmol·L-1 CaCl2,5 mmol·L-1 KCl,2 mmol·L-1 MES |
| MMg | 0.4 mol·L-1甘露醇Mannitol,15 mmol·L-1 MgCl2,4 mmol·L-1 MES |
| PEG 4000 | 20% PEG 4000,10 mmol·L-1 CaCl2,0.3 mol·L-1甘露醇Mannitol |
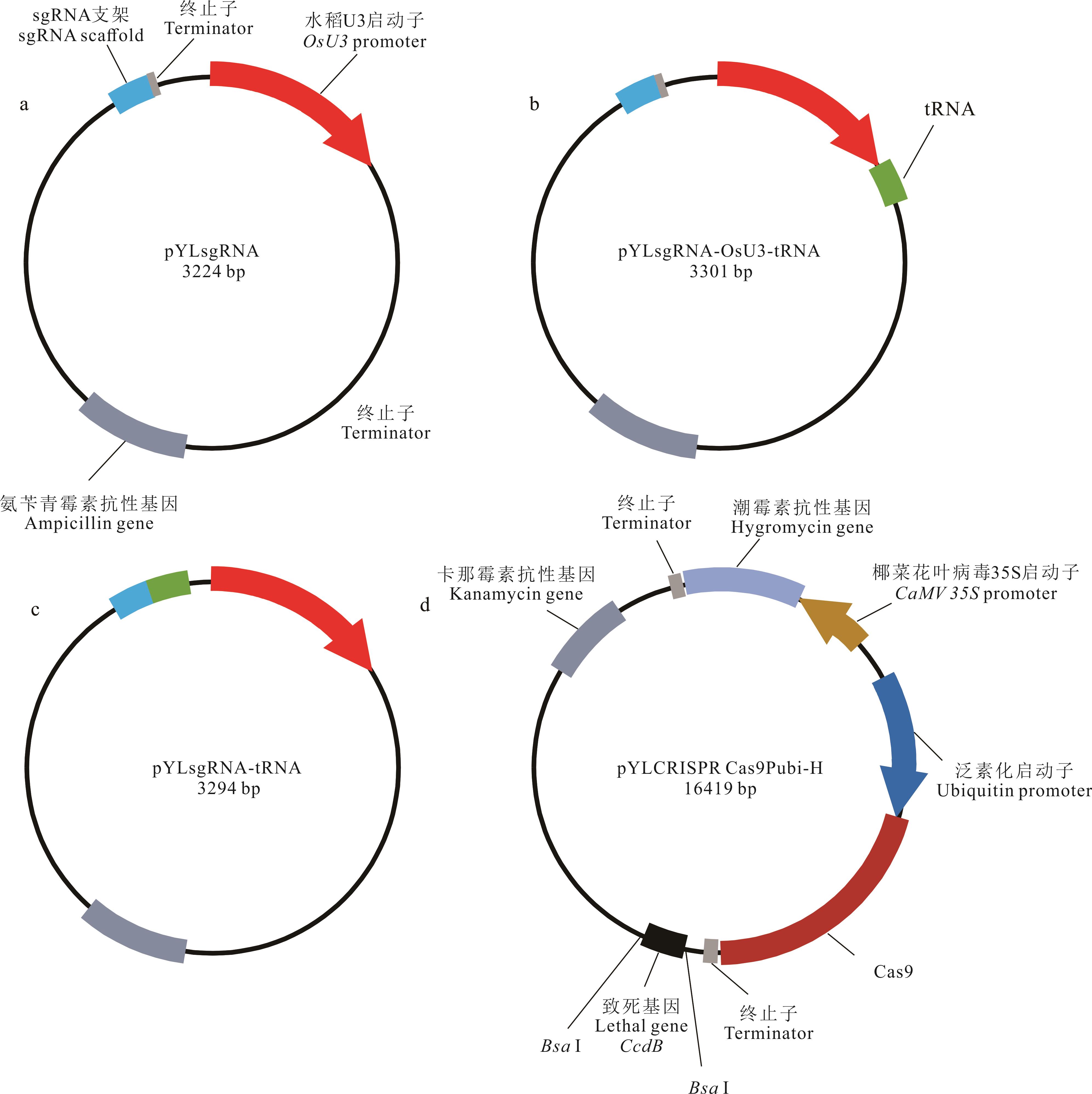
图2 构建tRNA-sgRNA多顺反子(PTG)表达盒的CRISPR/Cas9载体a: pYLsgRNA-OsU3原始载体pYLsgRNA-OsU3 original vector; b: pYLsgRNA-OsU3-tRNA(POT)载体pYLsgRNA-OsU3-tRNA (POT) vector; c: pYLsgRNA-tRNA(PT)载体pYLsgRNA-tRNA (PT) vector; d: pYLCRISPRCas9Pubi-H载体(Cas9双元载体) pYLCRISPRCas9Pubi-H vector (Cas9 binary vector).
Fig.2 CRISPR/Cas9 vectors for constructing the polycistronic tRNA-sgRNA (PTG) cassette
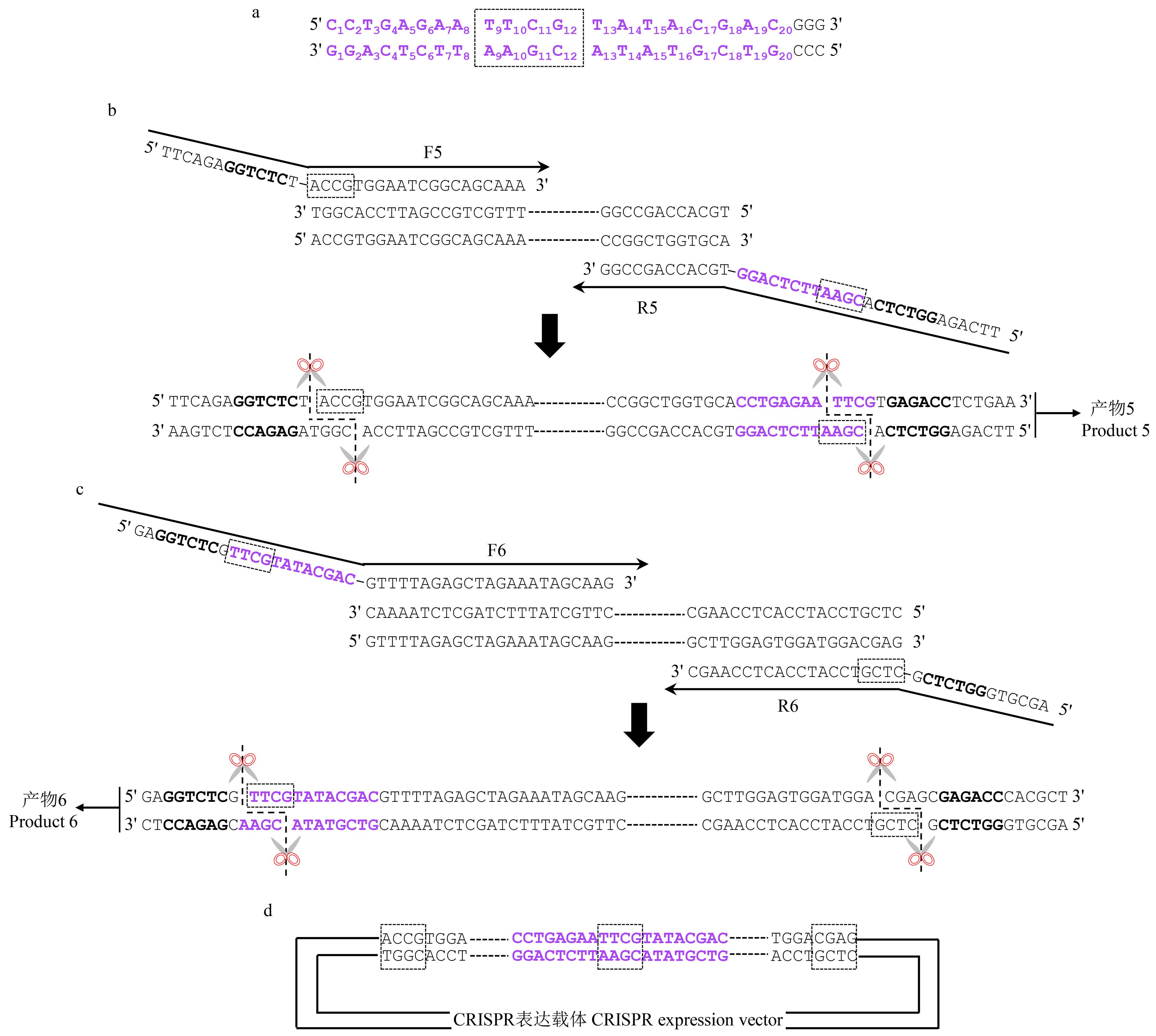
图4 Bsa I连接位点的选择及连接过程黑色加粗部分为Bsa Ⅰ识别序列,虚线方框为Bsa Ⅰ连接位点。The bold black parts are the Bsa Ⅰ recognition sequences, and the dotted boxs are the Bsa Ⅰ connection sites.
Fig.4 Selection of Bsa Ⅰ connection site and the connection process

图5 多年生黑麦草原生质体制备a: 制备的原生质体Isolated protoplasts; b~c: 原生质体的FDA染色观察Observation of FDA stained protoplasts; 原生质体经FDA染色后,b为一个视野中明场下的细胞总数量,c为相同视野中绿色荧光通路下的活细胞数量After the protoplasts were stained by FDA, Figure b shows the total number of cells under the bright light, and Figure c shows the number of viable cells under the green fluorescence light in the same field.
Fig.5 Perennial ryegrass protoplasts preparation
| 1 | Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012, 337: 816-821. |
| 2 | Le C, Ran F A, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-823. |
| 3 | Wiedenheft B, Sternberg S H, Doudna J A. RNA guided genetic silencing systems in bacteria and archaea. Nature, 2012, 482: 331-338. |
| 4 | Dicarlo J E, Norville J E, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research, 2013, 41(7): 4336-4343. |
| 5 | Jao L E, Wente S R, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(34): 13904-13909. |
| 6 | Gratz S J, Cummings A M, Nguyen J N, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics, 2013, 194(4): 1029-1035. |
| 7 | Cho S W, Kim S, Kim J M, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotechnology, 2013, 31(3): 230-232. |
| 8 | Jiang W Z, Zhou H B, Bi H H, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Research, 2013, 41(20): e188. |
| 9 | Makarova K S, Wolf Y I, Alkhnbashi O S, et al. An updated evolutionary classification of CRISPR-Cas systems. Nature Reviews Microbiology, 2015, 13(11): 722-736. |
| 10 | Jiang F G, Doudna J A. CRISPR-Cas9 structures and mechanisms. Annual Review of Biophysics, 2017, 46(1): 505-529. |
| 11 | Amitai G, Sorek R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nature Reviews Microbiology, 2016, 14(2): 67-76. |
| 12 | Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007, 315(5819): 1709-1712. |
| 13 | Li J F, Norville J E, Aach J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nature Biotechnology, 2013, 31(8): 688-691. |
| 14 | Ma X L, Zhang Q Y, Zhu Q L, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant, 2015, 8(8): 1274-1284. |
| 15 | Kabadi A M, Ousterout D G, Hilton I B, et al. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Research, 2014, 42(19): e147. |
| 16 | Cermak T, Curtin S J, Gil-Humanes J, et al. A multipurpose toolkit to enable advanced genome engineering in plants. The Plant Cell, 2017, 29(6): 1196-1217. |
| 17 | Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(11): 3570-3575. |
| 18 | White R J. Transcription by RNA polymerase Ⅲ: More complex than we thought. Nature Reviews Genetics, 2011, 12(7): 459-463. |
| 19 | Dieci G, Fiorino G, Castelnuovo M, et al. The expanding RNA polymerase Ⅲ transcriptome. Trends in Genetics, 2007, 23(12): 614-622. |
| 20 | Minkenberg B, Xie K, Yang Y. Discovery of rice essential genes by characterizing a CRISP-edited mutation of closely related rice MAP kinase genes. The Plant Journal, 2017, 89(3): 636-648. |
| 21 | Song Y L, Wang K Q, Wang S, et al. Physiological responses of three kinds of cool season turf grasses under continuous drought stress, heat stress and their interaction. Acta Agrestia Sinica, 2018, 26(3): 705-717. |
| 宋娅丽, 王克勤, 王莎, 等. 3种冷季型草坪草对持续干旱、高温及其互作的生理生态响应. 草地学报, 2018, 26(3): 705-717. | |
| 22 | Yu G H, Xie Z N, Chen W, et al. Knock down of non-yellow coloring 1-like gene or chlorophyll in application enhanced chlorophyll accumulation with antioxidant roles in suppressing heat-induced leaf senescence in perennial ryegrass. Journal of Experimental Botany, 2021, 73(1): 429-444. |
| 23 | Yu G H, Xie Z N, Zhang J, et al. NOL-mediated functional stay-green traits in perennial ryegrass (Lolium perenne L.) involving multifaceted molecular factors and metabolic pathways regulating leaf senescence. The Plant Journal, 2021, 106(5): 1219-1232. |
| 24 | Yu G, Cheng Q, Xie Z, et al. An efficient protocol for perennial ryegrass mesophyll protoplast isolation and transformation, and its application on interaction study between LpNOL and LpNYC1. Plant Methods, 2017, 13(1): 46. |
| 25 | Li J F, Zhang D D, Jen S. Targeted plant genome editing via the CRISPR/Cas9 technology. Methods in Molecular Biology, 2015, 1284: 239-255. |
| 26 | Hui L, Zhao M, He J, et al. A simple and reliable method for creating PCR-detectable mutants in Arabidopsis with the polycistronic tRNA-gRNA CRISPR/Cas9 system. Acta Physiologiae Plantarum, 2019, 41(10): 170-184. |
| 27 | Wang Z, Wang S, Li D, et al. Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnology Journal, 2018, 16(8): 1424-1433. |
| 28 | Engler C, Gruetzner R, Kandzia R, et al. Golden gate shuffling: A one-pot DNA shuffling method based on type Ⅱs restriction enzymes. PLoS One, 2009, 4(5): e5553. |
| 29 | Vidigal J A, Ventura A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nature Communications, 2015, 6(8083): 1-7. |
| 30 | Marraffini L A, Sontheimer E J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science, 2008, 322: 1843-1845. |
| 31 | Hale C R, Zhao P, Olson S, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell, 2009, 139(5): 945-956. |
| 32 | Garneau J E, Dupuis M E, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature, 2010, 468(7320): 67-71. |
| 33 | Karimi M, Meyer B D, Hilson P. Modular cloning in plant cells. Trends in Plant Science, 2005, 10(3): 103-105. |
| 34 | Zhang Y, Su J, Shan D, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods, 2011, 7(1): 30. |
| 35 | Qi W, Zhu T, Tian Z, et al. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnology, 2016, 16(1): 58. |
| 36 | Ren C, Liu Y, Guo Y, et al. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Horticulture Research, 2021, 8(1): 52-64. |
| [1] | 姚佳明, 何悦, 郝欢欢, 黄心如, 张敬, 徐彬. 多年生黑麦草LpPIL5基因特征分析及转录调控[J]. 草业学报, 2022, 31(9): 155-167. |
| [2] | 张晴, 邢静, 姚佳明, 殷庭超, 黄心如, 何悦, 张敬, 徐彬. 多年生黑麦草细胞分裂素信号通路B类ARR转录因子LpARR10的耐镉功能分析[J]. 草业学报, 2022, 31(5): 135-143. |
| [3] | 撖冬荣, 姚拓, 李海云, 黄书超, 杨琰珊, 高亚敏, 李昌宁, 张银翠. 微生物肥料与化肥减量配施对多年生黑麦草生长的影响[J]. 草业学报, 2022, 31(3): 136-143. |
| [4] | 赵利清, 彭向永, 刘俊祥, 毛金梅, 孙振元. GSH对铅胁迫下多年生黑麦草生长及光合生理的影响[J]. 草业学报, 2021, 30(9): 97-104. |
| [5] | 周瀚洋, 孙鹏越, 尉欣荣, 周雨, 张智伟, 高金柱, 赵东豪, 罗艺岚, 呼天明, 付娟娟. 琥珀酸黄杆菌缓解遮阴对多年生黑麦草胁迫的效应[J]. 草业学报, 2020, 29(6): 137-143. |
| [6] | 魏勇, 王晓瑜, 李应德, 段廷玉. AM真菌在白三叶-黑麦草体系中对抗逆信号的传导作用[J]. 草业学报, 2020, 29(4): 138-146. |
| [7] | 马碧花, 蔺伟虎, 高敏, 王兴迪, 田沛. 干旱胁迫下水杨酸和内生真菌对多年生黑麦草的影响[J]. 草业学报, 2020, 29(1): 135-144. |
| [8] | 王日明, 王志强, 向佐湘. γ-氨基丁酸对高温胁迫下黑麦草光合特性及碳水化合物代谢的影响[J]. 草业学报, 2019, 28(2): 168-178. |
| [9] | 李孟湛, 尹红菊, 李丁丁, 刘亚琪, 王锁民. 利用CRISPR/Cas9技术敲除拟南芥转录因子MYB 40的两种可变剪接体[J]. 草业学报, 2019, 28(1): 120-127. |
| [10] | 祁泽文, 孙鑫博, 樊波, 张雪, 袁建波, 韩烈保. PEG介导的柳枝稷叶肉细胞原生质体瞬时表达体系的建立[J]. 草业学报, 2017, 26(9): 113-120. |
| [11] | 包爱科, 白天惠, 赵天璇, 苏家豪. CRISPR/Cas9系统:基因组定点编辑技术及其在植物基因功能研究中的应用[J]. 草业学报, 2017, 26(7): 190-200. |
| [12] | 王沛, 崔彦农, 高丽, 王锁民. 盐生植物小花碱茅CYP86A基因的RNAi载体构建[J]. 草业学报, 2017, 26(6): 105-110. |
| [13] | 郭艳娥, 王晓瑜, 高萍, 段廷玉. 磷添加条件下摩西球囊霉与禾草内生真菌对多年生黑麦草生长的影响[J]. 草业学报, 2017, 26(12): 160-169. |
| [14] | 王日明, 熊兴耀. 高温胁迫对黑麦草生长及生理代谢的影响[J]. 草业学报, 2016, 25(8): 81-90. |
| [15] | 魏树强, 孙振元, 代小梅, 钱永强. 多年生黑麦草LpGCS基因克隆及其在烟草中的初步功能验证[J]. 草业学报, 2016, 25(4): 121-132. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||