

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2021, Vol. 30 ›› Issue (11): 144-156.DOI: 10.11686/cyxb2020429
• 研究论文 • 上一篇
收稿日期:2020-09-22
修回日期:2020-11-02
出版日期:2021-10-19
发布日期:2021-10-19
通讯作者:
张吉宇
作者简介:Corresponding author. E-mail: zhangjy@lzu.edu.cn基金资助:
Qian MA( ), Qi YAN, Zheng-she ZHANG, Fan WU, Ji-yu ZHANG(
), Qi YAN, Zheng-she ZHANG, Fan WU, Ji-yu ZHANG( )
)
Received:2020-09-22
Revised:2020-11-02
Online:2021-10-19
Published:2021-10-19
Contact:
Ji-yu ZHANG
摘要:
紫花苜蓿是全世界广泛栽培的优质牧草。木质素与紫花苜蓿抗性密切相关,但木质素难以被牲畜消化,严重影响了紫花苜蓿的营养价值,所以培育低木质素的紫花苜蓿具有重要意义。咖啡酰辅酶A氧甲基转移酶(CCoAOMT)是木质素合成途径中的关键酶,本研究利用生物信息学方法对紫花苜蓿基因组中的CCoAOMT基因家族成员进行鉴定,并对其基因结构、染色体定位、系统发育、基因表达等方面进行了研究。结果表明,在紫花苜蓿基因组中共鉴定到44个MsCCoAOMT基因,其中36个(82%)MsCCoAOMT基因都含有5个外显子,分布于16条染色体上,存在明显的串联重复现象;通过系统发育树分析,MsCCoAOMT基因家族可分为5类;通过MEME软件对MsCCoAOMT蛋白motif进行预测发现了10个比较保守的motif。qRT-PCR表达分析表明部分MsCCoAOMT基因的表达具有组织特异性。研究结果对紫花苜蓿低木质素遗传改良具有潜在的理论指导意义和实际参考价值。
马倩, 闫启, 张正社, 吴凡, 张吉宇. 紫花苜蓿CCoAOMT基因家族的鉴定、进化及表达分析[J]. 草业学报, 2021, 30(11): 144-156.
Qian MA, Qi YAN, Zheng-she ZHANG, Fan WU, Ji-yu ZHANG. Identification, evolution and expression analysis of the CCoAOMT family genes in Medicago sativa[J]. Acta Prataculturae Sinica, 2021, 30(11): 144-156.
基因名称 Gene name | 引物序列 Primer sequence (5'-3') | 基因名称 Gene name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| MsCCoAOMT1 | F: TGCTGATCCATTGGTAAGTGAC | MsCCoAOMT25 | F: CTATACCCGGTGATGAAGGAC |
| R: TTGTCATCCCATCTCCAATAGGT | R: CAAAGCCAATACCTTTCCATCAG | ||
| MsCCoAOMT10 | F: TTCATGGGCACTTCACCT | MsCCoAOMT32 | F: CTATACCCGCTGATGAAGGAC |
| R: GGCTGTAATCTTTCCATCATATGG | R: AGCCAACACCTTTCCATCCT | ||
| MsCCoAOMT12 | F: AAGCAAACTGAATCTGGAAGAC | MsCCoAOMT41 | F: GGAACCTTATGGCTACACCT |
| R: TTGTCATGATGTTCCATGGGT | R: AAGCCAATACCTTTCCATCAG | ||
| MsCCoAOMT17 | F: TGCCTTTGGATGAAGCAC | MsCCoAOMT43 | F: CACCCATGCTTGAAGGAG |
| R: CAATTCCTATTACCTTGCCATCAG | R: AGCTAACACCTTTCCATCCT | ||
| MsCCoAOMT21 | F: GGCTGGTCAACTAATCACTC | MsCCoAOMT44 | F: CTAATGGGCATACTCTTGAAGCT |
| R: GGCTGTAATCTTTCCATCGT | R: AGCTATGATCTTTCCATCATCAGG | ||
| MsCCoAOMT23 | F: GAGATGACACTATCACACCCT | MsActin | F: GACAATGGAACTGGAATGG |
| R: AAGCCAATACCTTTCCATCAG | R: CAATACCGTGCTCAATGG |
表1 qRT-PCR 引物信息
Table 1 Primers used for qRT-PCR analysis
基因名称 Gene name | 引物序列 Primer sequence (5'-3') | 基因名称 Gene name | 引物序列 Primer sequence (5'-3') |
|---|---|---|---|
| MsCCoAOMT1 | F: TGCTGATCCATTGGTAAGTGAC | MsCCoAOMT25 | F: CTATACCCGGTGATGAAGGAC |
| R: TTGTCATCCCATCTCCAATAGGT | R: CAAAGCCAATACCTTTCCATCAG | ||
| MsCCoAOMT10 | F: TTCATGGGCACTTCACCT | MsCCoAOMT32 | F: CTATACCCGCTGATGAAGGAC |
| R: GGCTGTAATCTTTCCATCATATGG | R: AGCCAACACCTTTCCATCCT | ||
| MsCCoAOMT12 | F: AAGCAAACTGAATCTGGAAGAC | MsCCoAOMT41 | F: GGAACCTTATGGCTACACCT |
| R: TTGTCATGATGTTCCATGGGT | R: AAGCCAATACCTTTCCATCAG | ||
| MsCCoAOMT17 | F: TGCCTTTGGATGAAGCAC | MsCCoAOMT43 | F: CACCCATGCTTGAAGGAG |
| R: CAATTCCTATTACCTTGCCATCAG | R: AGCTAACACCTTTCCATCCT | ||
| MsCCoAOMT21 | F: GGCTGGTCAACTAATCACTC | MsCCoAOMT44 | F: CTAATGGGCATACTCTTGAAGCT |
| R: GGCTGTAATCTTTCCATCGT | R: AGCTATGATCTTTCCATCATCAGG | ||
| MsCCoAOMT23 | F: GAGATGACACTATCACACCCT | MsActin | F: GACAATGGAACTGGAATGG |
| R: AAGCCAATACCTTTCCATCAG | R: CAATACCGTGCTCAATGG |
基因名称 Gene name | 基因座 Gene ID | 蛋白质大小 Protein length (aa) | 分子量MW (kDa) | 等电点 pI | 疏水性指数 GRAVY | 基因序列长度Length of intron and exon (bp) | 亚细胞定位Subcellular localization | 拟南芥同源基因 Homologous gene of Arabidopsis |
|---|---|---|---|---|---|---|---|---|
| MsCCoAOMT1 | MS.gene039184.t1 | 309 | 34.3566 | 8.54 | -0.123 | 5315 | Cytoplasm | AT3G62000.1 |
| MsCCoAOMT2 | MS.gene068675.t1 | 293 | 32.6967 | 8.58 | -0.083 | 3724 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT3 | MS.gene063440.t1 | 293 | 32.7298 | 8.58 | -0.061 | 3345 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT4 | MS.gene062840.t1 | 267 | 29.9507 | 8.58 | -0.080 | 7396 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT5 | MS.gene049025.t1 | 251 | 27.9613 | 5.51 | 0.020 | 1473 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT6 | MS.gene26463.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1302 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT7 | MS.gene06173.t1 | 235 | 26.1382 | 5.51 | -0.023 | 1495 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT8 | MS.gene058635.t1 | 133 | 14.8534 | 5.59 | 0.041 | 724 | N and C | AT1G67980.1 |
| MsCCoAOMT9 | MS.gene028389.t1 | 229 | 25.7237 | 5.14 | -0.114 | 2386 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT10 | MS.gene55886.t1 | 235 | 26.0962 | 5.51 | -0.043 | 1495 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT11 | MS.gene28756.t1 | 229 | 25.7418 | 5.23 | -0.097 | 1842 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT12 | MS.gene059423.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT13 | MS.gene053535.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT14 | MS.gene053539.t1 | 248 | 28.0291 | 5.41 | -0.280 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT15 | MS.gene053537.t1 | 182 | 20.5040 | 6.31 | -0.066 | 881 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT16 | MS.gene20157.t1 | 235 | 26.1222 | 5.51 | -0.023 | 1492 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT17 | MS.gene35069.t1 | 229 | 25.7377 | 5.14 | -0.112 | 2454 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT18 | MS.gene047856.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1322 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT19 | MS.gene064029.t1 | 238 | 26.8580 | 5.23 | -0.174 | 2072 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT20 | MS.gene030303.t1 | 238 | 26.8720 | 5.35 | -0.175 | 2087 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT21 | MS.gene75055.t1 | 238 | 26.9041 | 5.35 | -0.182 | 2042 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT22 | MS.gene66349.t1 | 238 | 26.8580 | 5.23 | -0.174 | 2072 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT23 | MS.gene44534.t1 | 238 | 26.7568 | 5.58 | -0.132 | 1866 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT24 | MS.gene44535.t1 | 568 | 63.5446 | 4.89 | -0.170 | 4423 | Chloroplast | AT2G23250.1 |
| MsCCoAOMT25 | MS.gene011784.t1 | 248 | 27.9000 | 5.15 | -0.173 | 3149 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT26 | MS.gene011803.t1 | 248 | 27.8408 | 4.99 | -0.198 | 3883 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT27 | MS.gene011804.t1 | 246 | 27.7320 | 5.50 | -0.130 | 1624 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT28 | MS.gene30425.t1 | 187 | 20.9131 | 5.22 | -0.100 | 2679 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT29 | MS.gene011778.t1 | 238 | 26.7828 | 5.48 | -0.124 | 2777 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT30 | MS.gene56811.t1 | 248 | 27.8449 | 5.06 | -0.158 | 4031 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT31 | MS.gene56793.t1 | 248 | 27.8689 | 4.99 | -0.185 | 3744 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT32 | MS.gene56792.t1 | 189 | 21.6996 | 5.65 | 0.286 | 1330 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT33 | MS.gene074442.t1 | 235 | 26.3723 | 5.52 | -0.197 | 3568 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT34 | MS.gene90469.t1 | 238 | 26.7828 | 5.48 | -0.124 | 1850 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT35 | MS.gene90479.t1 | 248 | 27.9000 | 5.15 | -0.173 | 2947 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT36 | MS.gene90498.t1 | 248 | 27.8127 | 4.91 | -0.182 | 3043 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT37 | MS.gene90499.t1 | 246 | 27.7240 | 5.50 | -0.108 | 1637 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT38 | MS.gene051092.t1 | 235 | 26.4173 | 5.30 | -0.186 | 3565 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT39 | MS.gene041940.t1 | 238 | 26.7828 | 5.48 | -0.124 | 2498 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT40 | MS.gene041950.t1 | 248 | 27.9071 | 5.16 | -0.160 | 2984 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT41 | MS.gene99216.t1 | 248 | 27.8408 | 4.99 | -0.195 | 3776 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT42 | MS.gene99215.t1 | 246 | 27.7351 | 5.50 | -0.034 | 1458 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT43 | MS.gene99213.t1 | 246 | 27.7460 | 5.50 | -0.129 | 1637 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT44 | MS.gene25100.t1 | 235 | 26.4394 | 5.55 | -0.185 | 3584 | Cytoplasm | AT4G26220.1 |
表2 紫花苜蓿CCoAOMT基因家族信息
Table 2 The information of CCoAOMT gene family in M. sativa
基因名称 Gene name | 基因座 Gene ID | 蛋白质大小 Protein length (aa) | 分子量MW (kDa) | 等电点 pI | 疏水性指数 GRAVY | 基因序列长度Length of intron and exon (bp) | 亚细胞定位Subcellular localization | 拟南芥同源基因 Homologous gene of Arabidopsis |
|---|---|---|---|---|---|---|---|---|
| MsCCoAOMT1 | MS.gene039184.t1 | 309 | 34.3566 | 8.54 | -0.123 | 5315 | Cytoplasm | AT3G62000.1 |
| MsCCoAOMT2 | MS.gene068675.t1 | 293 | 32.6967 | 8.58 | -0.083 | 3724 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT3 | MS.gene063440.t1 | 293 | 32.7298 | 8.58 | -0.061 | 3345 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT4 | MS.gene062840.t1 | 267 | 29.9507 | 8.58 | -0.080 | 7396 | Chloroplast | AT3G62000.1 |
| MsCCoAOMT5 | MS.gene049025.t1 | 251 | 27.9613 | 5.51 | 0.020 | 1473 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT6 | MS.gene26463.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1302 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT7 | MS.gene06173.t1 | 235 | 26.1382 | 5.51 | -0.023 | 1495 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT8 | MS.gene058635.t1 | 133 | 14.8534 | 5.59 | 0.041 | 724 | N and C | AT1G67980.1 |
| MsCCoAOMT9 | MS.gene028389.t1 | 229 | 25.7237 | 5.14 | -0.114 | 2386 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT10 | MS.gene55886.t1 | 235 | 26.0962 | 5.51 | -0.043 | 1495 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT11 | MS.gene28756.t1 | 229 | 25.7418 | 5.23 | -0.097 | 1842 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT12 | MS.gene059423.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT13 | MS.gene053535.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT14 | MS.gene053539.t1 | 248 | 28.0291 | 5.41 | -0.280 | 1303 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT15 | MS.gene053537.t1 | 182 | 20.5040 | 6.31 | -0.066 | 881 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT16 | MS.gene20157.t1 | 235 | 26.1222 | 5.51 | -0.023 | 1492 | Chloroplast | AT4G26220.1 |
| MsCCoAOMT17 | MS.gene35069.t1 | 229 | 25.7377 | 5.14 | -0.112 | 2454 | Chloroplast | AT1G67980.1 |
| MsCCoAOMT18 | MS.gene047856.t1 | 247 | 27.9992 | 5.55 | -0.308 | 1322 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT19 | MS.gene064029.t1 | 238 | 26.8580 | 5.23 | -0.174 | 2072 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT20 | MS.gene030303.t1 | 238 | 26.8720 | 5.35 | -0.175 | 2087 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT21 | MS.gene75055.t1 | 238 | 26.9041 | 5.35 | -0.182 | 2042 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT22 | MS.gene66349.t1 | 238 | 26.8580 | 5.23 | -0.174 | 2072 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT23 | MS.gene44534.t1 | 238 | 26.7568 | 5.58 | -0.132 | 1866 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT24 | MS.gene44535.t1 | 568 | 63.5446 | 4.89 | -0.170 | 4423 | Chloroplast | AT2G23250.1 |
| MsCCoAOMT25 | MS.gene011784.t1 | 248 | 27.9000 | 5.15 | -0.173 | 3149 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT26 | MS.gene011803.t1 | 248 | 27.8408 | 4.99 | -0.198 | 3883 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT27 | MS.gene011804.t1 | 246 | 27.7320 | 5.50 | -0.130 | 1624 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT28 | MS.gene30425.t1 | 187 | 20.9131 | 5.22 | -0.100 | 2679 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT29 | MS.gene011778.t1 | 238 | 26.7828 | 5.48 | -0.124 | 2777 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT30 | MS.gene56811.t1 | 248 | 27.8449 | 5.06 | -0.158 | 4031 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT31 | MS.gene56793.t1 | 248 | 27.8689 | 4.99 | -0.185 | 3744 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT32 | MS.gene56792.t1 | 189 | 21.6996 | 5.65 | 0.286 | 1330 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT33 | MS.gene074442.t1 | 235 | 26.3723 | 5.52 | -0.197 | 3568 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT34 | MS.gene90469.t1 | 238 | 26.7828 | 5.48 | -0.124 | 1850 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT35 | MS.gene90479.t1 | 248 | 27.9000 | 5.15 | -0.173 | 2947 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT36 | MS.gene90498.t1 | 248 | 27.8127 | 4.91 | -0.182 | 3043 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT37 | MS.gene90499.t1 | 246 | 27.7240 | 5.50 | -0.108 | 1637 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT38 | MS.gene051092.t1 | 235 | 26.4173 | 5.30 | -0.186 | 3565 | Cytoplasm | AT4G26220.1 |
| MsCCoAOMT39 | MS.gene041940.t1 | 238 | 26.7828 | 5.48 | -0.124 | 2498 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT40 | MS.gene041950.t1 | 248 | 27.9071 | 5.16 | -0.160 | 2984 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT41 | MS.gene99216.t1 | 248 | 27.8408 | 4.99 | -0.195 | 3776 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT42 | MS.gene99215.t1 | 246 | 27.7351 | 5.50 | -0.034 | 1458 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT43 | MS.gene99213.t1 | 246 | 27.7460 | 5.50 | -0.129 | 1637 | Cytoplasm | AT4G34050.1 |
| MsCCoAOMT44 | MS.gene25100.t1 | 235 | 26.4394 | 5.55 | -0.185 | 3584 | Cytoplasm | AT4G26220.1 |

图1 紫花苜蓿CCoAOMT基因的染色体定位标尺表示染色体长度,黑色箭头表示基因的转录方向,红色字体标注串联重复基因。The scale indicates the chromosome length, the black arrow indicates the transcription direction of the gene, and the red font marks the tandem repeat gene.
Fig.1 Location of MsCCoAOMT genes on chromosome
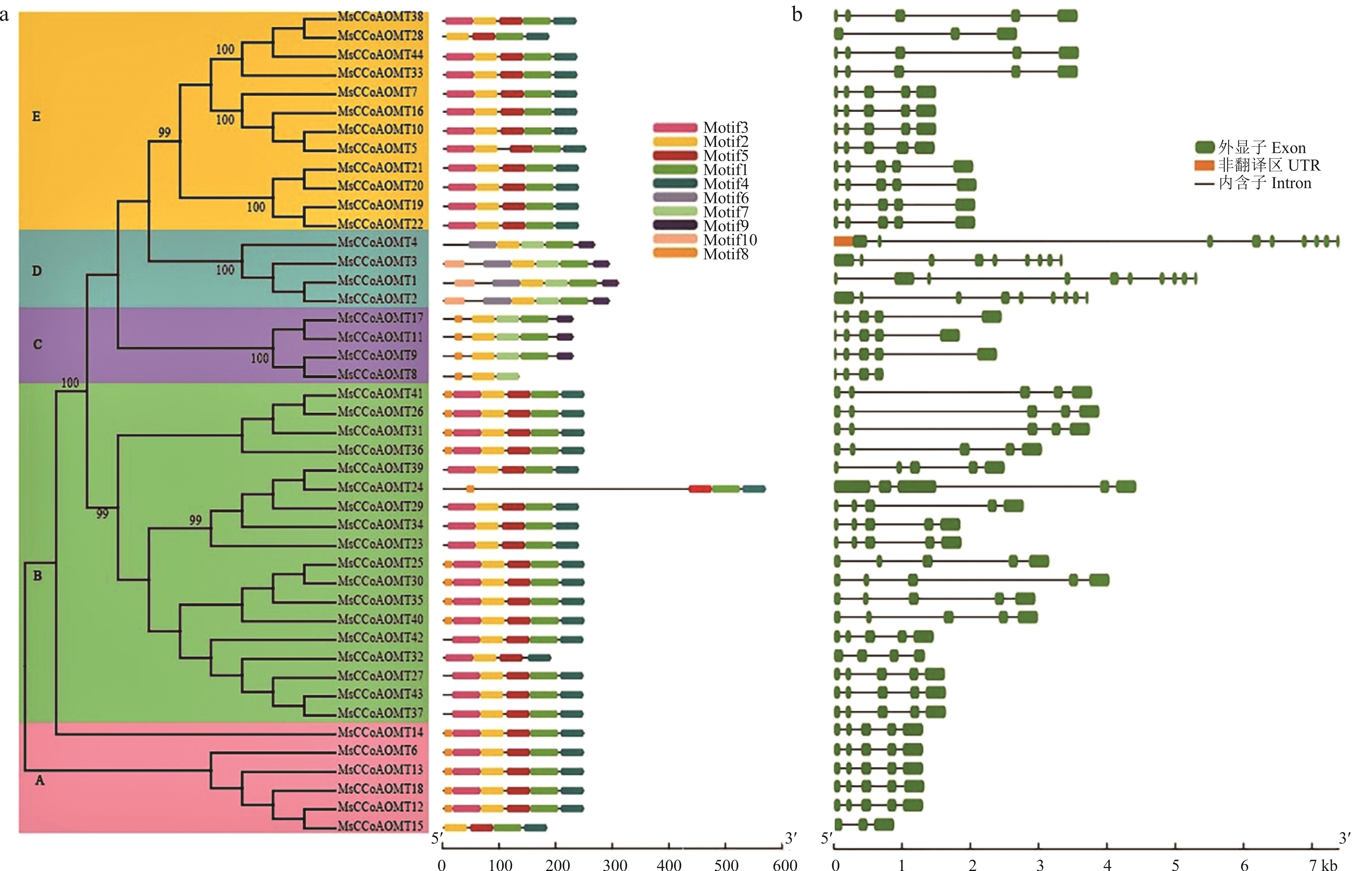
图2 紫花苜蓿CCoAOMT基因家族的保守基序及基因结构分析a. MsCCoAOMT基因家族的保守基序;b. MsCCoAOMT基因家族的基因结构,绿色框、黑色线条分别代表外显子和内含子;5个不同的背景色表示五大类。a. Conserved motifs of the MsCCoAOMT family; b. Gene structure of the MsCCoAOMT family. The green boxes and black lines represent exon and intron, respectively; Class A, B, C, D, and E are displayed in different background colors.
Fig.2 Analysis of conserved motifs and gene structure of the MsCCoAOMT gene family
基因名称 Gene name | 元件名称Element name | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
基本核心元件 Basic core components | 光信号感知顺式作用元件Optical signal sensing cis-acting element | 激素响应元件 Hormone response element | 生物应激反应元件 Biological stress response element | 蛋白质结合元件 Protein binding element | 组织发育元件 Tissue development element | |||||||||||||||||||||||||||
| X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 | X13 | X14 | X15 | X16 | X17 | X18 | X19 | X20 | X21 | X22 | X23 | X24 | X25 | X26 | X27 | X28 | X29 | X30 | X31 | X32 | |
| Ms1 | 46 | 24 | 2 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms2 | 34 | 22 | 1 | 0 | 1 | 3 | 1 | 1 | 2 | 1 | 3 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms3 | 43 | 14 | 3 | 2 | 3 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms4 | 33 | 11 | 0 | 0 | 2 | 2 | 2 | 0 | 2 | 0 | 3 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Ms5 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms6 | 32 | 47 | 1 | 0 | 2 | 4 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms7 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms8 | 41 | 51 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms9 | 39 | 56 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms10 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms11 | 38 | 54 | 0 | 0 | 4 | 1 | 3 | 2 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| Ms12 | 31 | 43 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms13 | 33 | 47 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms14 | 23 | 75 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms15 | 43 | 36 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ms16 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms17 | 39 | 56 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms18 | 32 | 48 | 1 | 0 | 2 | 3 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms19 | 44 | 60 | 0 | 0 | 3 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms20 | 39 | 43 | 0 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ms21 | 43 | 60 | 0 | 0 | 2 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms22 | 44 | 61 | 0 | 0 | 2 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms23 | 41 | 25 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms24 | 38 | 33 | 0 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms25 | 39 | 60 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms26 | 29 | 21 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 |
| Ms27 | 39 | 53 | 0 | 0 | 4 | 2 | 1 | 1 | 1 | 0 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms28 | 53 | 46 | 1 | 1 | 5 | 4 | 3 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Ms29 | 40 | 27 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms30 | 37 | 66 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms31 | 39 | 56 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms32 | 47 | 43 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms33 | 46 | 28 | 0 | 0 | 2 | 3 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| Ms34 | 42 | 29 | 1 | 1 | 4 | 2 | 4 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms35 | 42 | 61 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms36 | 33 | 64 | 0 | 1 | 3 | 1 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| Ms37 | 44 | 67 | 0 | 1 | 5 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms38 | 46 | 31 | 0 | 0 | 2 | 3 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ms39 | 42 | 26 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms40 | 29 | 43 | 1 | 1 | 7 | 2 | 2 | 1 | 1 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Ms41 | 40 | 59 | 0 | 1 | 3 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms42 | 44 | 61 | 0 | 1 | 4 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms43 | 44 | 65 | 0 | 1 | 4 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms44 | 48 | 23 | 0 | 1 | 3 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
表3 启动子序列顺式作用元件分析
Table 3 Analysis of promoter sequence cis-acting
基因名称 Gene name | 元件名称Element name | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
基本核心元件 Basic core components | 光信号感知顺式作用元件Optical signal sensing cis-acting element | 激素响应元件 Hormone response element | 生物应激反应元件 Biological stress response element | 蛋白质结合元件 Protein binding element | 组织发育元件 Tissue development element | |||||||||||||||||||||||||||
| X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | X12 | X13 | X14 | X15 | X16 | X17 | X18 | X19 | X20 | X21 | X22 | X23 | X24 | X25 | X26 | X27 | X28 | X29 | X30 | X31 | X32 | |
| Ms1 | 46 | 24 | 2 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms2 | 34 | 22 | 1 | 0 | 1 | 3 | 1 | 1 | 2 | 1 | 3 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms3 | 43 | 14 | 3 | 2 | 3 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms4 | 33 | 11 | 0 | 0 | 2 | 2 | 2 | 0 | 2 | 0 | 3 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Ms5 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms6 | 32 | 47 | 1 | 0 | 2 | 4 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms7 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms8 | 41 | 51 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms9 | 39 | 56 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms10 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms11 | 38 | 54 | 0 | 0 | 4 | 1 | 3 | 2 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| Ms12 | 31 | 43 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms13 | 33 | 47 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms14 | 23 | 75 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms15 | 43 | 36 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Ms16 | 35 | 39 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms17 | 39 | 56 | 1 | 0 | 2 | 4 | 3 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms18 | 32 | 48 | 1 | 0 | 2 | 3 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Ms19 | 44 | 60 | 0 | 0 | 3 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms20 | 39 | 43 | 0 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ms21 | 43 | 60 | 0 | 0 | 2 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms22 | 44 | 61 | 0 | 0 | 2 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms23 | 41 | 25 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms24 | 38 | 33 | 0 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Ms25 | 39 | 60 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms26 | 29 | 21 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 |
| Ms27 | 39 | 53 | 0 | 0 | 4 | 2 | 1 | 1 | 1 | 0 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms28 | 53 | 46 | 1 | 1 | 5 | 4 | 3 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Ms29 | 40 | 27 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms30 | 37 | 66 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms31 | 39 | 56 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms32 | 47 | 43 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms33 | 46 | 28 | 0 | 0 | 2 | 3 | 2 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| Ms34 | 42 | 29 | 1 | 1 | 4 | 2 | 4 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms35 | 42 | 61 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 |
| Ms36 | 33 | 64 | 0 | 1 | 3 | 1 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| Ms37 | 44 | 67 | 0 | 1 | 5 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms38 | 46 | 31 | 0 | 0 | 2 | 3 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Ms39 | 42 | 26 | 1 | 1 | 4 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Ms40 | 29 | 43 | 1 | 1 | 7 | 2 | 2 | 1 | 1 | 0 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Ms41 | 40 | 59 | 0 | 1 | 3 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ms42 | 44 | 61 | 0 | 1 | 4 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms43 | 44 | 65 | 0 | 1 | 4 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ms44 | 48 | 23 | 0 | 1 | 3 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |

图5 不同生长发育阶段紫花苜蓿‘中苜1号’不同组织中MsCCoAOMT基因相对表达量A1:幼苗期叶;A2:幼苗期茎;A3:营养期叶;A4:营养期茎;A5:花期花;A6:花期茎;A7:花期叶。A1: Leaves of seedling stage; A2: Stems of seedling stage; A3: Leaves of vegetative stage; A4: Stems of vegetative stage; A5: Florets of florescence; A6: Stems of florescence; A7: Leaves of florescence.
Fig.5 Relative expression of MsCCoAOMT gene in different tissues of M. sativa ‘Zhongmu No.1’ at different growth and development stages
| 1 | Li Y F, Cheng J H, Tian C Y, et al. Effects of sodium diacetate on the quality, nutrient composition and protein molecular structure of alfalfa silage. Acta Prataculturae Sinica, 2020, 29(2): 163-171. |
| 李艳芬, 程金花, 田川尧, 等. 双乙酸钠对苜蓿青贮品质、营养成分及蛋白分子结构的影响. 草业学报, 2020, 29(2): 163-171. | |
| 2 | Cheng Q M, Ge G T, Sa D W, et al. Transcription analyses provide insights into differences in nutritional quality among different alfalfa varieties. Acta Prataculturae Sinica, 2019, 28(10): 199-208. |
| 成启明, 格根图, 撒多文, 等. 不同品种紫花苜蓿转录组分析及营养品质差异的探讨. 草业学报, 2019, 28(10): 199-208. | |
| 3 | Zhang X Y, Guo A P, He L K, et al. Advances in study of lignin biosynthesis and its genetic mainpulation. Molecular Plant Breeding, 2006, 4(3): 431-437. |
| 章霄云, 郭安平, 贺立卡, 等. 木质素生物合成及其基因调控的研究进展. 分子植物育种, 2006, 4(3): 431-437. | |
| 4 | Hu K, Yan X F, Li D, et al. Genetic improvement of perennial ryegrass with low lignin content by silencing genes of CCR and CAD. Acta Prataculturae Sinica, 2013, 22(5): 72-83. |
| 胡可, 严雪锋, 栗丹, 等. 沉默CCR和CAD基因培育低木质素含量转基因多年生黑麦草. 草业学报, 2013, 22(5): 72-83. | |
| 5 | Pang S L, Ong S, Lee H, et al. Isolation and characterization of CCoAOMT in interspecific hybrid of Acacia auriculiformis×Acacia mangium——a key gene in lignin biosynthesis. Genetics and Molecular Research, 2014, 13(3): 7217-7238. |
| 6 | Yao L M, Hu X Q, Zhou F, et al. Antisense CCoAOMT gene regulates lignin biosynthesis in Betula platyphylla. Bulletin of Botanical Research, 2019, 39(1): 123-130. |
| 姚连梅, 胡晓晴, 周菲, 等. 白桦反义CCoAOMT基因调控木质素生物合成. 植物研究, 2019, 39(1): 123-130. | |
| 7 | Wu J Z, Qian C, Liu Z W, et al. De novo transcriptomic analysis for lignin synthesis in Cenchrus purpureus using RNA-seq. Acta Prataculturae Sinica, 2019, 28(1): 150-161. |
| 吴娟子, 钱晨, 刘智微, 等. 基于转录组测序分析象草木质素合成的研究. 草业学报, 2019, 28(1): 150-161. | |
| 8 | Zhu S Q, Xu D B, Wang Y X, et al. Cloning and expression characteristics analysis of caffeoyl-CoA O-methyltransferase gene in Oenanthe javanica. Journal of Nanjing Agricultural University, 2019, 42(1): 51-58. |
| 朱胜琪, 徐德宝, 王永鑫, 等. 水芹咖啡酰辅酶A-O-甲基转移酶基因的克隆与表达特性分析. 南京农业大学学报, 2019, 42(1): 51-58. | |
| 9 | Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Biology, 2003, 54(1): 519-546. |
| 10 | Schmitt D, Pakusch A E, Matern U. Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. Journal of Biological Chemistry, 1991, 266(26): 17416-17423. |
| 11 | Ye Z H, Kneusel R E, Matern U, et al. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell, 1994, 6(10): 1427-1439. |
| 12 | Do C T, Pollet B, Thévenin J, et al. Both caffeoyl coenzyme a 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta, 2007, 226: 1117-1129. |
| 13 | Huang C Q, Liu G D, Guo A P. Lignin biosynthesis regulated by antisense CCoAOMT gene in tobacco. Journal of Anhui Agricultural Sciences, 2008, 36(19): 8026-8027. |
| 黄春琼, 刘国道, 郭安平. 反义CCoAOMT基因调控烟草木质素的生物合成. 安徽农业科学, 2008, 36(19): 8026-8027. | |
| 14 | Pinçon G, Maury S, Hoffmann L, et al. Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry, 2001, 57(7): 1167-1176. |
| 15 | Guo D J, Chen F, Inoue K, et al. Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell, 2001, 13(1): 73-88. |
| 16 | Liu H R, Zhao H Y, Wei J H, et al. Effects of repression of CCoAOMT expression on lignin biosynthesis in tobacco. Scientia Agricultura Sinica, 2002, 35(8): 921-924. |
| 刘惠荣, 赵华燕, 魏建华, 等. 抑制CCoAOMT表达对烟草木质素生物合成的影响. 中国农业科学, 2002, 35(8): 921-924. | |
| 17 | Li X Y, Chen W J, Zhao Y, et al. Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genetics and Molecular Biology, 2013, 36(4): 540-546. |
| 18 | Wang Z J, Ge Q, Chen C, et al. Function analysis of caffeoyl-CoA O-methyltransferase for biosynthesis of lignin and phenolic acid in Salvia miltiorrhiza. Applied Biochemistry and Biotechnology, 2017, 181(2): 562-572. |
| 19 | Tetreault H M, Scully E D, Gries T, et al. Overexpression of the Sorghum bicolorSbCCoAOMT alters cell wall associated hydroxycinnamoyl groups. PLoS One, 2018, 13(10): e0204153. |
| 20 | Raes J, Rohde A, Christensen J H, et al. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiology, 2003, 133(3): 1051-1071. |
| 21 | Martz F, Maury S, Pinçon G, et al. cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyltransferase, a lignin biosynthetic enzyme. Plant Molecular Biology, 1998, 36(3): 427-437. |
| 22 | Zhao H Y, Shen Q X, Lv S Y, et al. Expression characteristics of cafeyl-coenzyme A-O-methyltransferase gene (CCoAOMT) in rice. Chinese Science Bulletin, 2004, 49(14): 1390-1394. |
| 赵华燕, 沈庆喜, 吕世友, 等. 水稻咖啡酰辅酶A-O-甲基转移酶基因(CCoAOMT)表达特性分析. 科学通报, 2004, 49(14): 1390-1394. | |
| 23 | Chen H T, Zeng Y, Yang Y Z, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nature Communications, 2020, 11(1): 2494. |
| 24 | Luo Y, Liu Y B, Dong Y X, et al. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment. Journal of Plant Physiology, 2009, 166(4): 385-394. |
| 25 | Zhang J Y, Wang Y R, Nan Z B. Relative and absolute quantification expression analysis of CsSAMDC gene as a case. China Biotechnology, 2009, 29(8): 86-91. |
| 张吉宇, 王彦荣, 南志标. 相对定量和绝对定量—以CsSAMDC基因表达分析为例. 中国生物工程杂志, 2009, 29(8): 86-91. | |
| 26 | Huang C Q, Liu G D, Bai C J. Application advances of genetic engineering in pasture breeding. Acta Agrestia Sinica, 2013, 21(3): 413-419. |
| 黄春琼, 刘国道, 白昌军. 基因工程在牧草育种中的应用进展. 草地学报, 2013, 21(3): 413-419. | |
| 27 | Wang D Y, Gao F Y, Sun W, et al. Cloning and expression analysis of CCoAOMT gene in fruit of ‘Dangshansuli’. Journal of Nanjing Agricultural University, 2015, 38(1): 33-40. |
| 王丹阳, 高付永, 孙炜, 等. ‘砀山酥梨’果实CCoAOMT基因的克隆与表达分析. 南京农业大学学报, 2015, 38(1): 33-40. | |
| 28 | Gu Z J, Zhang D Q, Huang Q Y. Transgenic modification on pulp plants by key genes regulating lignin biosynthesis. Journal of Central South University of Forestry & Technology, 2010, 30(3): 67-74. |
| 谷振军, 张党权, 黄青云. 木质素合成关键酶基因与造纸植物转基因改良应用研究. 中南林业科技大学学报, 2010, 30(3): 67-74. | |
| 29 | Gao Y, Chen X B, Zhang Z Y. Advances in research on lignin biosynthesis and its molecular regulation. Biotechnology Bulletin, 2007(2): 47-51. |
| 高原, 陈信波, 张志扬. 木质素生物合成途径及其基因调控的研究进展. 生物技术通报, 2007(2): 47-51. | |
| 30 | Bian J H, Liu Z Y, Li Z Y, et al. Advances in alfalfa improvement using genetic engineering. Pratacultural Science, 2020, 37(1): 139-155. |
| 边佳辉, 刘自扬, 李宗英, 等. 紫花苜蓿基因工程改良研究进展. 草业科学, 2020, 37(1): 139-155. | |
| 31 | Barros J, Temple S, Dixon R A. Development and commercialization of reduced lignin alfalfa. Current Opinion in Biotechnology, 2019, 56: 48-54. |
| 32 | Lu J, Zhao H Y, Wei J H, et al. Lignin reduction in transgenic poplars by expressing antisense CCoAOMT gene. Progress in Natural Science, 2004, 14(12): 1060-1063. |
| [1] | 赵颖, 辛夏青, 魏小红. 一氧化氮对干旱胁迫下紫花苜蓿氮代谢的影响[J]. 草业学报, 2021, 30(9): 86-96. |
| [2] | 汪雪, 刘晓静, 赵雅姣, 王静. 根系分隔方式下紫花苜蓿/燕麦间作氮素利用及种间互馈特征研究[J]. 草业学报, 2021, 30(8): 73-85. |
| [3] | 古丽娜扎尔·艾力null, 陶海宁, 王自奎, 沈禹颖. 基于APSIM模型的黄土旱塬区苜蓿——小麦轮作系统深层土壤水分及水分利用效率研究[J]. 草业学报, 2021, 30(7): 22-33. |
| [4] | 周倩倩, 张亚见, 张静, 殷涂童, 盛下放, 何琳燕. 产硫化氢细菌的筛选及阻控苜蓿吸收铅和改良土壤的作用[J]. 草业学报, 2021, 30(7): 44-52. |
| [5] | 臧真凤, 白婕, 刘丛, 昝看卓, 龙明秀, 何树斌. 紫花苜蓿形态和生理指标响应干旱胁迫的品种特异性[J]. 草业学报, 2021, 30(6): 73-81. |
| [6] | 谢展, 穆麟, 张志飞, 陈桂华, 刘洋, 高帅, 魏仲珊. 乳酸菌或有机酸盐与尿素复配添加对紫花苜蓿混合青贮的影响[J]. 草业学报, 2021, 30(5): 165-173. |
| [7] | 王吉祥, 宫焕宇, 屠祥建, 郭侲洐, 赵嘉楠, 沈健, 栗振义, 孙娟. 耐亚磷酸盐紫花苜蓿品种筛选及评价指标的鉴定[J]. 草业学报, 2021, 30(5): 186-199. |
| [8] | 张小芳, 魏小红, 刘放, 朱雪妹. PEG胁迫下紫花苜蓿幼苗内源激素对NO的响应[J]. 草业学报, 2021, 30(4): 160-169. |
| [9] | 候怡谣, 李霄, 龙瑞才, 杨青川, 康俊梅, 郭长虹. 过量表达紫花苜蓿MsHB7基因对拟南芥耐旱性的影响[J]. 草业学报, 2021, 30(4): 170-179. |
| [10] | 马欣, 罗珠珠, 张耀全, 刘家鹤, 牛伊宁, 蔡立群. 黄土高原雨养区不同种植年限紫花苜蓿土壤细菌群落特征与生态功能预测[J]. 草业学报, 2021, 30(3): 54-67. |
| [11] | 畅志鹏, 孙莹莹, 李佳阳, 龚春梅. 柠条CkCAD基因的克隆转化及其抗旱功能分析[J]. 草业学报, 2021, 30(3): 68-80. |
| [12] | 沙栢平, 谢应忠, 高雪芹, 蔡伟, 伏兵哲. 地下滴灌水肥耦合对紫花苜蓿草产量及品质的影响[J]. 草业学报, 2021, 30(2): 102-114. |
| [13] | 李倩, 李晓霞, 程丽琴, 陈双燕, 齐冬梅, 杨伟光, 高利军, 新巴音, 刘公社. 羊草LcCBF6基因的表达特性和功能研究[J]. 草业学报, 2021, 30(10): 105-115. |
| [14] | 王如月, 袁世力, 文武武, 周鹏, 安渊. 磷对铝胁迫紫花苜蓿幼苗根系生长和生理特征的影响[J]. 草业学报, 2021, 30(10): 53-62. |
| [15] | 李振松, 万里强, 李硕, 李向林. 苜蓿根系构型及生理特性对干旱复水的响应[J]. 草业学报, 2021, 30(1): 189-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||