

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (4): 186-198.DOI: 10.11686/cyxb2023193
• 研究论文 • 上一篇
李硕1( ), 李培英1,2,3(
), 李培英1,2,3( ), 孙宗玖1,2,3, 李雯1
), 孙宗玖1,2,3, 李雯1
收稿日期:2023-06-09
修回日期:2023-08-28
出版日期:2024-04-20
发布日期:2024-01-15
通讯作者:
李培英
作者简介:E-mail: nmlpy_1234@sina.com基金资助:
Shuo LI1( ), Pei-ying LI1,2,3(
), Pei-ying LI1,2,3( ), Zong-jiu SUN1,2,3, Wen LI1
), Zong-jiu SUN1,2,3, Wen LI1
Received:2023-06-09
Revised:2023-08-28
Online:2024-04-20
Published:2024-01-15
Contact:
Pei-ying LI
摘要:
根系是狗牙根响应干旱胁迫的重要器官,其响应不同程度干旱的分子机制有待进一步明确。以抗旱(C138)和敏旱(C32)2个狗牙根基因型为材料,对其正常灌溉(土壤相对含水量为田间持水量的80%~90%)、中度干旱(50%~60%)及重度干旱(20%~30%)下的根系进行取样,基于Illumina高通量测序平台进行转录组测序,共获得43581条单基因,有33025条得到注释。结果表明,与正常灌溉相比,中度及重度干旱下C138共7537个差异基因表达,其中上调1164个,下调6373个;C32有6731个差异基因表达,其中上调4304个,下调2427个。Top20 GO富集分析发现,中度干旱下C138主要富集于氧化还原酶活性、碳水化合物代谢和纤维素结合等,重度干旱下C138主要富集于磷酸代谢及蛋白磷酸化等,C32主要富集于过氧化物酶活性、氧化应激反应等;KEGG Pathway富集分析发现中度干旱下C138主要富集在谷胱甘肽代谢、苯丙基类生物合成和氮代谢途径;C32主要富集在氧化磷酸化、三羧酸循环(TCA)、核糖体和碳代谢途径;重度干旱胁迫下C138主要富集在谷胱甘肽代谢、花生四烯酸代谢、核糖体酶途径;C32主要富集在谷胱甘肽代谢、苯丙基类生物合成途径。总体而言,狗牙根中度干旱胁迫基因表达响应主要与氧化反应有关,重度干旱胁迫主要与Ca2+通路、脱落酸(ABA)信号通路和谷胱甘肽代谢有关,因而谷胱甘肽代谢可能是狗牙根响应干旱的主要KEGG通路。且与C32相比,C138响应干旱胁迫时有更多的钙依赖性蛋白激酶相关基因表达,因而抗旱性能更强。综上,谷胱甘肽代谢和MYB转录因子相关基因可能是狗牙根抗旱的关键,可作为其干旱胁迫基因研究的首选。
李硕, 李培英, 孙宗玖, 李雯. 基于转录组测序的狗牙根抗旱根系关键代谢途径分析[J]. 草业学报, 2024, 33(4): 186-198.
Shuo LI, Pei-ying LI, Zong-jiu SUN, Wen LI. Transcriptome analysis-based bermudagrass root RNA sequencing data under drought stress[J]. Acta Prataculturae Sinica, 2024, 33(4): 186-198.
| 编号Code | 采集地点Collection location | 抗旱类型Drought resistance type |
|---|---|---|
| C32 | 新疆吐鲁番市托克逊县Tuokexun County, Turpan City, Xinjiang Autonomous Region | 敏旱型Drought sensitive |
| C138 | 新疆喀什疏勒县Shule County, Kashgar City, Xinjiang Autonomous Region | 抗旱型Drought resistant |
表 1 材料具体信息
Table 1 Materials details
| 编号Code | 采集地点Collection location | 抗旱类型Drought resistance type |
|---|---|---|
| C32 | 新疆吐鲁番市托克逊县Tuokexun County, Turpan City, Xinjiang Autonomous Region | 敏旱型Drought sensitive |
| C138 | 新疆喀什疏勒县Shule County, Kashgar City, Xinjiang Autonomous Region | 抗旱型Drought resistant |
| 基因ID Gene ID | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| Actin | GCTCAACCCCAAGGCTAACAG | AGCGTATCCCTCGTAGATG |
| c458080.graph_c0 | CGGTGCTGGTTGGGTAGTCTG | AAATGGCGGCAATAGCGAAAGC |
| c452636.graph_c0 | CCCTGCCCAACGACGAAGAG | CATTGGCTTGGCGTGCTTGAG |
| c422082.graph_c1 | GCCCATCACCGCCAATCCG | CAAACGCATTAGCAACGCAACG |
| c448662.graph_c0 | GTGAGGTCCAGGCGGTGTTG | CGGCGAGTCGGTGTTCCTTG |
| c443527.graph_c0 | TGCCCGTTGACCTGTCTTTCTC | AGCACTTCCTCCGCCATTTCG |
| c460219.graph_c1 | GCGGCGAGCAATGATGTTACAG | AGATGGCTGGCGTTCTTGGC |
表 2 差异基因引物序列
Table 2 Differential gene primer sequences
| 基因ID Gene ID | 正向引物Forward primer (5′-3′) | 反向引物Reverse primer (5′-3′) |
|---|---|---|
| Actin | GCTCAACCCCAAGGCTAACAG | AGCGTATCCCTCGTAGATG |
| c458080.graph_c0 | CGGTGCTGGTTGGGTAGTCTG | AAATGGCGGCAATAGCGAAAGC |
| c452636.graph_c0 | CCCTGCCCAACGACGAAGAG | CATTGGCTTGGCGTGCTTGAG |
| c422082.graph_c1 | GCCCATCACCGCCAATCCG | CAAACGCATTAGCAACGCAACG |
| c448662.graph_c0 | GTGAGGTCCAGGCGGTGTTG | CGGCGAGTCGGTGTTCCTTG |
| c443527.graph_c0 | TGCCCGTTGACCTGTCTTTCTC | AGCACTTCCTCCGCCATTTCG |
| c460219.graph_c1 | GCGGCGAGCAATGATGTTACAG | AGATGGCTGGCGTTCTTGGC |
样品 Sample | 过滤后序列长度Clean datas length (bp) | GC含量 GC content (%) | 碱基质量>20 Q20 (%) | 碱基质量>30 Q30 (%) | 样品 Sample | 过滤后序列长度Clean datas length (bp) | GC含量 GC content (%) | 碱基质量>20 Q20 (%) | 碱基质量>30 Q30 (%) |
|---|---|---|---|---|---|---|---|---|---|
| CR138-1 | 5751481130 | 52.51 | 97.67 | 93.70 | MR32-1 | 7662111708 | 52.97 | 97.70 | 93.80 |
| CR138-2 | 6361023526 | 52.89 | 97.81 | 94.00 | MR32-2 | 7405754772 | 52.73 | 97.77 | 93.94 |
| CR138-3 | 5963989470 | 53.19 | 97.69 | 93.80 | MR32-3 | 7729678484 | 53.38 | 97.79 | 94.03 |
| CR32-1 | 8694639522 | 51.96 | 98.29 | 95.02 | SR138-1 | 6680468588 | 52.06 | 97.67 | 93.72 |
| CR32-2 | 7119356674 | 52.58 | 98.21 | 94.90 | SR138-2 | 6788905702 | 52.18 | 97.37 | 92.94 |
| CR32-3 | 6517861366 | 51.93 | 97.87 | 94.05 | SR138-3 | 5945423404 | 52.11 | 97.86 | 94.16 |
| MR138-1 | 9409664642 | 52.57 | 97.75 | 93.90 | SR32-1 | 6205146120 | 52.31 | 97.94 | 94.31 |
| MR138-2 | 6314881976 | 52.21 | 97.70 | 93.72 | SR32-2 | 6469804754 | 52.11 | 97.86 | 94.08 |
| MR138-3 | 6298108628 | 52.30 | 97.67 | 93.69 | SR32-3 | 6935579000 | 51.64 | 97.81 | 94.00 |
表3 转录组数据质控
Table 3 Quality control of the transcriptomic data
样品 Sample | 过滤后序列长度Clean datas length (bp) | GC含量 GC content (%) | 碱基质量>20 Q20 (%) | 碱基质量>30 Q30 (%) | 样品 Sample | 过滤后序列长度Clean datas length (bp) | GC含量 GC content (%) | 碱基质量>20 Q20 (%) | 碱基质量>30 Q30 (%) |
|---|---|---|---|---|---|---|---|---|---|
| CR138-1 | 5751481130 | 52.51 | 97.67 | 93.70 | MR32-1 | 7662111708 | 52.97 | 97.70 | 93.80 |
| CR138-2 | 6361023526 | 52.89 | 97.81 | 94.00 | MR32-2 | 7405754772 | 52.73 | 97.77 | 93.94 |
| CR138-3 | 5963989470 | 53.19 | 97.69 | 93.80 | MR32-3 | 7729678484 | 53.38 | 97.79 | 94.03 |
| CR32-1 | 8694639522 | 51.96 | 98.29 | 95.02 | SR138-1 | 6680468588 | 52.06 | 97.67 | 93.72 |
| CR32-2 | 7119356674 | 52.58 | 98.21 | 94.90 | SR138-2 | 6788905702 | 52.18 | 97.37 | 92.94 |
| CR32-3 | 6517861366 | 51.93 | 97.87 | 94.05 | SR138-3 | 5945423404 | 52.11 | 97.86 | 94.16 |
| MR138-1 | 9409664642 | 52.57 | 97.75 | 93.90 | SR32-1 | 6205146120 | 52.31 | 97.94 | 94.31 |
| MR138-2 | 6314881976 | 52.21 | 97.70 | 93.72 | SR32-2 | 6469804754 | 52.11 | 97.86 | 94.08 |
| MR138-3 | 6298108628 | 52.30 | 97.67 | 93.69 | SR32-3 | 6935579000 | 51.64 | 97.81 | 94.00 |
| 长度Length | 转录本Transcript | 单基因Unigene |
|---|---|---|
| 300~500 bp | 18373 | 11200 |
| 500~1000 bp | 28421 | 11052 |
| 1000~2000 bp | 35521 | 10442 |
| >2000 bp | 33675 | 10887 |
| 总数Total | 115990 | 43581 |
| 总长度Total length (nt) | 189838956 | 63773469 |
| N50 (bp) | 2321 | 2315 |
| 平均长度Mean length (nt) | 1636.68 | 1463.33 |
表4 转录组数据组装统计
Table 4 Data assembly of the RNA-Seq results
| 长度Length | 转录本Transcript | 单基因Unigene |
|---|---|---|
| 300~500 bp | 18373 | 11200 |
| 500~1000 bp | 28421 | 11052 |
| 1000~2000 bp | 35521 | 10442 |
| >2000 bp | 33675 | 10887 |
| 总数Total | 115990 | 43581 |
| 总长度Total length (nt) | 189838956 | 63773469 |
| N50 (bp) | 2321 | 2315 |
| 平均长度Mean length (nt) | 1636.68 | 1463.33 |
功能数据库 Annotation database | 被注释Unigene数 Annotated Unigene number | 300≤ 长度Length <1000 | 长度 Length ≥1000 |
|---|---|---|---|
| COG | 9436 | 2955 | 6481 |
| GO | 24135 | 8902 | 15233 |
| KEGG | 19248 | 6593 | 12655 |
| KOG | 16086 | 5556 | 10530 |
| Pfam | 22934 | 7878 | 15056 |
| Swissprot | 17945 | 5537 | 12408 |
| TrEMBL | 28867 | 10495 | 18372 |
| eggNOG | 22752 | 7551 | 15201 |
| NR | 32195 | 13237 | 18958 |
| All | 33025 | 13855 | 19170 |
表5 Unigenes注释统计
Table 5 Statistics of Unigenes annotated
功能数据库 Annotation database | 被注释Unigene数 Annotated Unigene number | 300≤ 长度Length <1000 | 长度 Length ≥1000 |
|---|---|---|---|
| COG | 9436 | 2955 | 6481 |
| GO | 24135 | 8902 | 15233 |
| KEGG | 19248 | 6593 | 12655 |
| KOG | 16086 | 5556 | 10530 |
| Pfam | 22934 | 7878 | 15056 |
| Swissprot | 17945 | 5537 | 12408 |
| TrEMBL | 28867 | 10495 | 18372 |
| eggNOG | 22752 | 7551 | 15201 |
| NR | 32195 | 13237 | 18958 |
| All | 33025 | 13855 | 19170 |

图 2 各材料及不同处理间差异基因GO富集图柱形图右边数字表示富集于该项上的基因数目The number on the right of the histogram indicates the amounts of genes enriched in this items.
Fig.2 The top 20 GO terms of DEGs in different materials and treatments

图4 干旱胁迫下谷胱甘肽代谢通路中差异表达基因变化A: C138中与谷胱甘肽途径相关的DEGs表达热图Heat map of DEGs expression related to glutathione pathway in C138; B,C: C138谷胱甘肽途径DEGs表达热图补充C138 Glutathione pathway DEGs expression heat map supplement; D: C32中与谷胱甘肽途径相关的DEGs表达热图Heat map of DEGs expression related to glutathione pathway in C32.
Fig.4 Changes of DEGs in the metabolic pathway of glutathione under drought stress
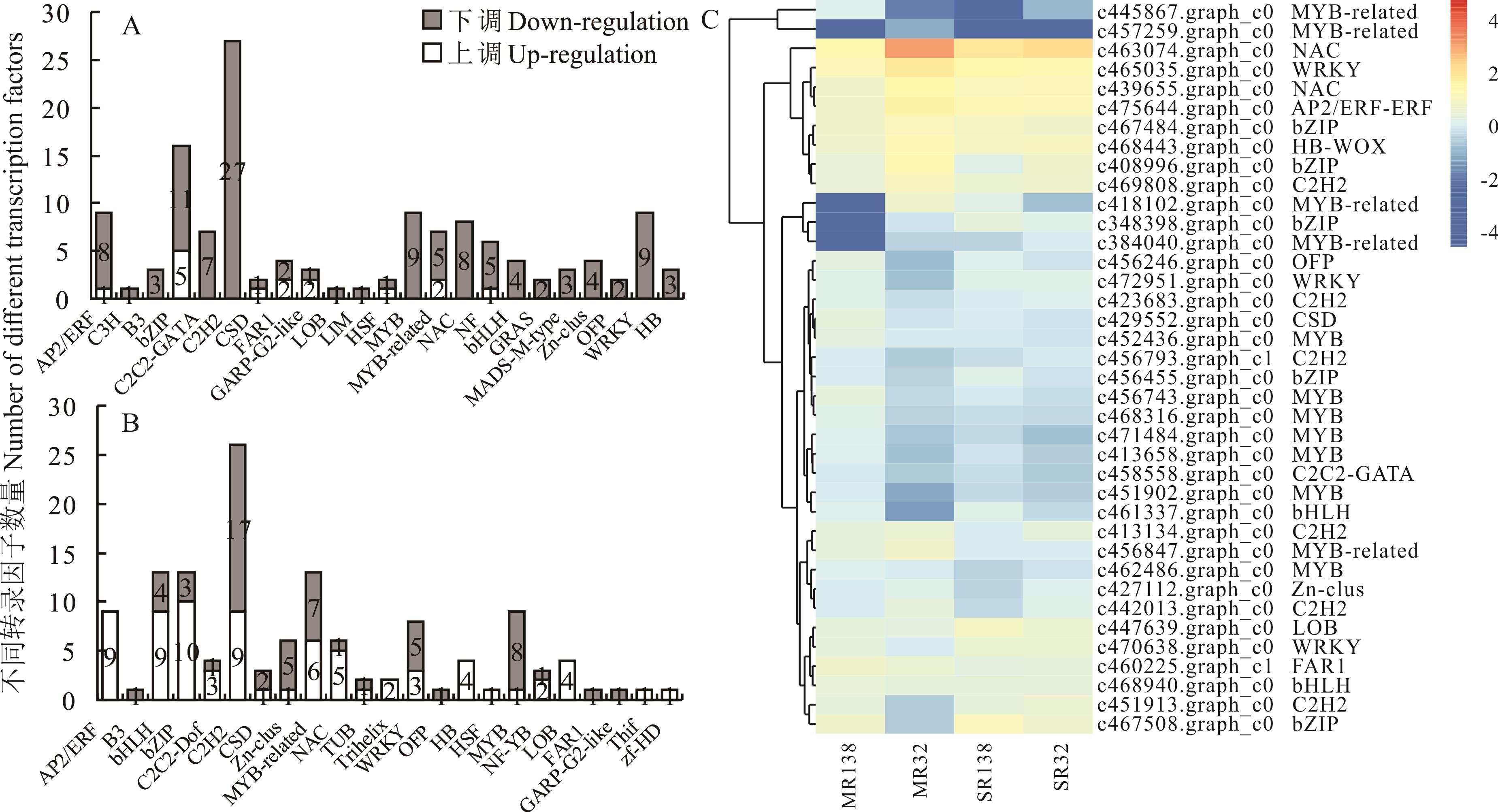
图5 干旱胁迫下狗牙根根系差异表达转录因子的分布A: C138根系Histogram of C138 root transcription factors; B: C32根系Histogram of C32 root transcription factors; C: 共表达差异基因DEG-TF的分级聚类Hierarchical clustering of DEG-TF co-expression genes.
Fig.5 The histograms represent the number of up- and down-regulated transcription factors
| 1 | Eriyagama N, Smakhtin V, Gamage N. Mapping drought patterns and impacts: A global perspective. Colombo Sri Lanka: International Water Management Institute (IWMI) (Research Reports 133), 2009. |
| 2 | Huang B. Mechanisms and strategies for improving drought resistance in turfgrass. Acta Horticulturae, 2008, 783(6): 221-232. |
| 3 | Bian S, Jiang Y. Reactive oxygen species, antioxidant enzyme activities and gene expression patterns in leaves and roots of Kentucky bluegrass in response to drought stress and recovery. Scientia Horticulturae, 2009, 120(2): 264-270. |
| 4 | Merewitz E B, Gianfagna T, Huang B. Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. Journal of Experimental Botany, 2011, 62(15): 5311-5333. |
| 5 | Cao Y J, Wei Q, Liao Y, et al. Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Reports, 2009, 28(4): 579-588. |
| 6 | Shi H T, Wang Y P, Cheng Z M, et al. Analysis of natural variation in bermudagrass (Cynodon dactylon) reveals physiological responses underlying drought tolerance. PLoS One, 2012, 7(12): 12-20. |
| 7 | Lu S, Chen C, Wang Z, et al. Physiological responses of somaclonal variants of triploid bermudagrass (Cynodon transvaalensis×Cynodon dactylon) to drought stress. Plant Cell Reports, 2009, 28(3): 517-526. |
| 8 | Zhou Y, Lambrides C J, Shu F K. Drought resistance of bermudagrass (Cynodon spp.) ecotypes collected from different climatic zones. Environmental and Experimental Botany, 2013, 41(5): 505-519. |
| 9 | Shabier A, Abudureyimu A, Younusi Q. Comparisons of drought resistance of six Xinjiang bermudagrass varieties.Journal of Xinjiang Agricultural University, 2008(2): 17-21. |
| 阿力木·沙比尔, 阿不来提·阿不都热依木, 齐曼·尤努斯. 6份新疆狗牙根抗旱性比较. 新疆农业大学学报, 2008(2): 17-21. | |
| 10 | Duan M M. Evaluation of drought resistance and physiological response of Xingjiang wild bermudagrass germplasm. Urumqi: Xinjiang Agricultural University, 2016. |
| 段敏敏. 新疆野生狗牙根种质抗旱性鉴定及抗旱生理的研究. 乌鲁木齐: 新疆农业大学, 2016. | |
| 11 | Zeng L S, Li P Y. Evaluation on drought resistance of 10 bermudagrass (Cynodon dactylon) germplasms from Xinjiang.Chinese Journal of Grassland, 2019, 41(3): 22-29. |
| 曾令霜, 李培英. 10份新疆狗牙根种质抗旱性评价. 中国草地学报, 2019, 41(3): 22-29. | |
| 12 | Zhou Y, Lambrides C J, Fukai S. Drought resistance of C4 grasses under field conditions: genetic variation among a large number of bermudagrass (Cynodon spp.) ecotypes collected from different climatic zones. Journal of Agronomy Crop Science, 2013, 199(4): 253-263. |
| 13 | Zeng L S, Li P Y, Sun X F, et al. A multi-trait evaluation of drought resistance of bermudagrass (Cynodon dactylon) germplasm from different habitats in Xinjiang province. Acta Prataculturae Sinica, 2020, 29(8): 155-169. |
| 曾令霜, 李培英, 孙晓梵, 等. 新疆不同生境狗牙根种质抗旱性综合评价. 草业学报, 2020, 29(8): 155-169. | |
| 14 | Zhang Y L, Yu Q K, Li W, et al. Aboveground and belowground phenotypic characteristics of Cynodon dactylon lines differing in drought resistance and endogenous hormone response to drought stress. Acta Prataculturae Sinica, 2023, 32(3): 163-178. |
| 张一龙, 喻启坤, 李雯, 等. 不同抗旱性狗牙根地上地下表型特征及内源激素对干旱胁迫的响应. 草业学报, 2023, 32(3): 163-178. | |
| 15 | Anders S, Huber W. Differential expression analysis for sequence count data. Nature Precedings, 2010, 11(10): R106. |
| 16 | Hou Z H, Yin J L, Lu Y F, et al. Transcriptomic analysis reveals the temporal and spatial changes in physiological process and gene expression in common buckwheat (Fagopyrum esculentum Moench) grown under drought stress. Agronomy, 2019, 9(10): 17-26. |
| 17 | Shao A, Wang W, Fan S G, et al. Comprehensive transcriptional analysis reveals salt stress-regulated key pathways, hub genes and time-specific responsive gene categories in common bermudagrass (Cynodon dactylon (L.) Pers.) roots. BMC Plant Biology, 2021, 21(1): 18-26. |
| 18 | Guan S J, Wang N, Xu R R, et al. Photosynthesis,antioxidant enzyme activity, and transcriptome sequencing analyses of Glyeyrrrhiza uralensis seedlings in response to drought stress. Pratacultural Science, 2021, 38(11): 2176-2190. |
| 关思静, 王楠, 徐蓉蓉, 等. 甘草幼苗响应干旱胁迫的光合、抗氧化特性及转录组分析.草业科学, 2021, 38(11): 2176-2190. | |
| 19 | Zhang J J, Li J J, Nian H J. The role of calcium/calmodulin signaling pathways in the stresses: Progress in researches. Chinese Journal of Microecology, 2013, 25(7): 858-860. |
| 张晶晶, 李金金, 年洪娟. 钙/钙调素信号途径在胁迫中的作用研究进展.中国微生态学杂志, 2013, 25(7): 858-860. | |
| 20 | He L. Relationship between 2C serine/threonine protein phosphatase activity and drought tolerance in maize. Chengdu: Sichuan Agricultural University, 2008. |
| 何亮. 玉米2C型丝氨酸/苏氨酸蛋白磷酸酶活性与耐旱性的关系.成都: 四川农业大学, 2008. | |
| 21 | Meskiene I, Baudouin E, Schweighofer A, et al. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. Journal of Biological Chemistry, 2003, 278(21): 18945-18952. |
| 22 | Danquah A, De Zélicourt A, Colcombet J, et al. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnology Advances, 2014, 32(1): 40-52. |
| 23 | Xiong L, Schumaker K S, Zhu J K. Cell signaling during cold, drought, and salt stress. Plant Cell, 2002, 14(5): 165-183. |
| 24 | Lin Y F, Li W, Dai L Y. Research progress of antioxidant enzymes functioning in plant drought resistant process. Crop Research, 2015, 29(3): 326-330. |
| 林宇丰, 李魏, 戴良英. 抗氧化酶在植物抗旱过程中的功能研究进展.作物研究, 2015, 29(3): 326-330. | |
| 25 | Foyer C H, Noctor G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiology, 2011, 155(1): 2-18. |
| 26 | Song G F, Fan W L, Wang J J, et al.Cloning and characterization of drought-stress responsive gene GhGR in Gossypium hirsutum L. Scientia Agricultura Sinica, 2012, 45(8): 1644-1652. |
| 宋贵方, 樊伟丽, 王俊娟, 等. 陆地棉干旱胁迫响应基因GhGR的克隆及特征分析. 中国农业科学, 2012, 45(8): 1644-1652. | |
| 27 | Shan C J, Han R L, Liang Z S.Responses to drought stress of the biosynthetic and recycling metabolism of glutathione and ascorbate in Agropyron cristatum leaves on the Loess Plateau of China.Chinese Journal of Plant Ecology, 2011, 35(6): 653-662. |
| 单长卷, 韩蕊莲, 梁宗锁.黄土高原冰草叶片抗坏血酸和谷胱甘肽合成及循环代谢对干旱胁迫的生理响应. 植物生态学报, 2011, 35(6): 653-662. | |
| 28 | Kim C, Lemke C, Paterson A H. Functional dissection of drought-responsive gene expression patterns in Cynodon dactylon L. Plant Molecular Biology, 2009, 70(2): 1-16. |
| 29 | Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 2003, 15(1): 63-78. |
| 30 | Guo H, Wang Y, Wang L, et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnology Journal, 2017, 15(1): 107-121. |
| 31 | Hu L X, Li H Y, Chen L, et al. RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genomics, 2015, 16: 1-12. |
| 32 | Yuan X, Huang P, Wang R, et al. A zinc finger transcriptional repressor confers pleiotropic effects on rice growth and drought tolerance by down-regulating stress-responsive genes. Plant Cell Physiology, 2018, 59(10): 2129-2142. |
| [1] | 鲍根生, 李媛, 冯晓云, 张鹏, 孟思宇. 高寒区氮添加和间作种植互作对燕麦和豌豆根系构型影响的研究[J]. 草业学报, 2024, 33(3): 73-84. |
| [2] | 汪雪, 刘晓静, 王静, 吴勇, 童长春. 连续间作下的紫花苜蓿/燕麦根系与碳氮代谢特性研究[J]. 草业学报, 2024, 33(3): 85-96. |
| [3] | 曾兵, 尚盼盼, 沈秉娜, 王胤晨, 屈明好, 袁扬, 毕磊, 杨兴云, 李文文, 周晓丽, 郑玉倩, 郭文强, 冯彦龙, 曾兵. 淹水胁迫下鸭茅根系基因差异表达及相关通路分析[J]. 草业学报, 2024, 33(2): 93-111. |
| [4] | 姜瑛, 张辉红, 魏畅, 徐正阳, 赵颖, 刘芳, 李鸽子, 张雪海, 柳海涛. 外源褪黑素对干旱胁迫下玉米幼苗根系发育及生理生化特性的影响[J]. 草业学报, 2023, 32(9): 143-159. |
| [5] | 王宝强, 马文静, 王贤, 朱晓林, 赵颖, 魏小红. 一氧化氮对干旱胁迫下紫花苜蓿幼苗次生代谢产物的影响[J]. 草业学报, 2023, 32(8): 141-151. |
| [6] | 魏艳, 刘有斌, 刘枭宏, 谌芸, 颜哲豪, 都艺芝. 紫色土区拉巴豆和紫花苜蓿根-土复合体抗剪性能研究[J]. 草业学报, 2023, 32(8): 82-90. |
| [7] | 杨瑞杰, 何淑勤, 周树峰, 杨晶月, 金钰宪, 郑子成. 杂交粱草生长期土壤抗冲性变化特征及其根系调控效应[J]. 草业学报, 2023, 32(7): 149-159. |
| [8] | 廖小琴, 王长庭, 刘丹, 唐国, 毛军. 氮磷配施对高寒草甸植物根系特征的影响[J]. 草业学报, 2023, 32(7): 160-174. |
| [9] | 张一龙, 李雯, 喻启坤, 李培英, 孙宗玖. 狗牙根叶与根氮代谢对不同干旱胁迫的响应机制[J]. 草业学报, 2023, 32(7): 175-187. |
| [10] | 张浩, 胡海英, 李惠霞, 贺海明, 马霜, 马风华, 宋柯辰. 荒漠草原优势植物牛枝子对干旱胁迫的生理响应与转录组分析[J]. 草业学报, 2023, 32(7): 188-205. |
| [11] | 梁佳, 胡朝阳, 谢志明, 马刘峰, 陈芸, 方志刚. 外源褪黑素缓解甜高粱幼苗干旱胁迫的生理效应[J]. 草业学报, 2023, 32(7): 206-215. |
| [12] | 陈晓明, 韩东英, 宋桂龙. 砷(As)胁迫对海滨雀稗As吸收特征及根系形态影响[J]. 草业学报, 2023, 32(6): 112-119. |
| [13] | 崔婷, 王勇, 马晖玲. 外源IAA作用下草地早熟禾中调控Cd长距离运输的关键基因表达及其代谢通路分析[J]. 草业学报, 2023, 32(6): 146-156. |
| [14] | 杨瑞鑫, 李勇, 蔡小芳, 韩铖星, 郭艳丽. 不同物理形态的开食料对羔羊瘤胃转录组的影响[J]. 草业学报, 2023, 32(5): 159-170. |
| [15] | 李艳鹏, 魏娜, 翟庆妍, 李杭, 张吉宇, 刘文献. 全基因组水平白花草木樨TCP基因家族的鉴定及在干旱胁迫下表达模式分析[J]. 草业学报, 2023, 32(4): 101-111. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||