

ISSN 1004-5759 CN 62-1105/S


草业学报 ›› 2024, Vol. 33 ›› Issue (11): 84-105.DOI: 10.11686/cyxb2023492
白世俊1,2,3( ), 李军乔1,2,3(
), 李军乔1,2,3( ), 刘欣1,2,3, 吕博文1,2,3
), 刘欣1,2,3, 吕博文1,2,3
收稿日期:2023-12-19
修回日期:2024-03-21
出版日期:2024-11-20
发布日期:2024-09-09
通讯作者:
李军乔
作者简介:E-mail: jqlily2002@126.com基金资助:
Shi-jun BAI1,2,3( ), Jun-qiao LI1,2,3(
), Jun-qiao LI1,2,3( ), Xin LIU1,2,3, Bo-wen LYU1,2,3
), Xin LIU1,2,3, Bo-wen LYU1,2,3
Received:2023-12-19
Revised:2024-03-21
Online:2024-11-20
Published:2024-09-09
Contact:
Jun-qiao LI
摘要:
青藏高原的隆升以及第四纪冰期气候的循环波动,对于青藏高原及其周边地区动植物的分布和遗传结构具有很大的影响。蕨麻是青藏高原极富营养、药用和生态价值的特有植物资源,蕨麻和鹅绒委陵菜的分类关系及分布在学术界存在争议,分子谱系地理学研究将为蕨麻遗传多样性形成机制及推断该物种迁移演化历史提供依据。以蕨麻为研究对象,对采集的30个居群的810个个体进行了叶绿体trnL-trnF序列和核基因ITS测序,揭示遗传变异在居群内和居群间的分布格局,结合群体遗传学和系统发生学,分析该物种的遗传结构与历史事件之间的关联,揭示物种及物种内不同种群形成现有分布格局的历史原因和演化过程。主要结论有:1)蕨麻具有较高水平的遗传多样性。cpDNA trnL-trnF片段共检测到40种单倍型,16个为共享单倍型,占比40%,24个为居群特有单倍型,占比60%,遗传多样性h=0.7078,单倍型多样性Hd=0.8217,核苷酸多样性π=0.010641,总遗传多样性HT=0.849;nrDNA ITS片段共检测到128种单倍型,共享单倍型42种,占比32.8%,居群特有单倍型86种,占比67.2%,遗传多样性h=0.7633,单倍型多样性Hd=0.8168,核苷酸多样性π=0.003584,总遗传多样性HT=0.844。2)居群内的遗传多样性大于居群间的遗传多样性。序列分析结果为居群内和居群间的遗传多样性都很高(cpDNA trnL-trnF:HT=0.849,居群内平均遗传多样性HS=0.640;nrDNA ITS序列:HT=0.844,HS=0.763,HT均大于HS)。蕨麻居群分为3个组:青海高原组、横断山脉组和藏南谷地组。蕨麻的遗传变异主要来源于居群内部。3)蕨麻种群具有明显的谱系地理结构。cpDNA trnL-trnF序列和nrDNA ITS遗传多态性分析及地理分布模式检验,cpDNA trnL-trnF序列:遗传分化系数GST=0.246,NST=0.417,nrDNA ITS序列:GST=0.096,NST=0.522,NST均显著大于GST(P<0.001),表明蕨麻所有居群单倍型存在显著的谱系地理结构,两种方法的结果高度一致。分子变异分析(AMOVA)表明,大部分遗传变异(59.69%)存在于居群内部,居群间分化水平很高(FST=0.40313)。4)共享单倍型和特有单倍型均由古老单倍型衍生而来。cpDNA trnL-trnF和nrDNA序列中央连接网状图呈以共享单倍型M4和H9位于中心,M1、M3和H2、H10、H11、H12位于主干位置的星状结构,其余共享单倍型和特有单倍型均由这些古老单倍型衍生而来,两者结果一致。5)蕨麻种群大小和范围发生过大规模扩张。利用cpDNA trnL-trnF和nrDNA序列进行歧点分析,前者歧点分布呈单峰曲线,表明近期群体大小和范围有大规模扩张发生;后者的歧点分布呈双峰曲线,反映基因谱系的高度复杂性,但Tajima’s D,Fu and Li’s D和Fu and Li’s F均为负值,且结果显著,且离差平方和(SSD)和扩张评估指数(HRag)的统计检验不显著,表明蕨麻居群近期有扩张的可能。6)蕨麻存在3个冰期避难所,即东喜马拉雅区域、青藏高原东南边缘及横断山脉区域。
白世俊, 李军乔, 刘欣, 吕博文. 青藏高原蕨麻的分子谱系地理学研究[J]. 草业学报, 2024, 33(11): 84-105.
Shi-jun BAI, Jun-qiao LI, Xin LIU, Bo-wen LYU. Phylogeography of Potentilla anserina distributed across the Qinghai-Tibet Plateau[J]. Acta Prataculturae Sinica, 2024, 33(11): 84-105.
编号 Code | 采集地 Locality | 经度 Longitude (°, E) | 纬度 Latitude (°, N) | 海拔 Altitude (m) | 采集数 Collection number (No.) |
|---|---|---|---|---|---|
| P1 | 青海化隆Hualong, Qinghai | 102.070718 | 36.178380 | 2696 | 22 |
| P2 | 青海泽库Zeku, Qinghai | 101.480690 | 35.012647 | 3625 | 28 |
| P3 | 甘肃碌曲Luqu, Gansu | 102.763742 | 34.818995 | 2972 | 24 |
| P4 | 西藏察雅Chaya, Tibet | 97.057961 | 30.661889 | 4366 | 15 |
| P5 | 西藏八宿Basu, Tibet | 96.791225 | 29.469206 | 3913 | 11 |
| P6 | 西藏左贡Zuogong, Tibet | 97.915733 | 29.674744 | 3997 | 17 |
| P7 | 西藏芒康Mangkang, Tibet | 98.585117 | 29.600331 | 3812 | 25 |
| P8 | 云南香格里拉Shangri-La, Yunnan | 99.666918 | 27.872883 | 3267 | 20 |
| P9 | 西藏安多Anduo, Tibet | 91.514772 | 31.407749 | 4598 | 25 |
| P10 | 西藏比如Biru, Tibet | 93.417949 | 31.286509 | 3909 | 26 |
| P11 | 西藏浪卡子Langkazi, Tibet | 90.237586 | 28.594107 | 4443 | 28 |
| P12 | 西藏措美Cuomei, Tibet | 91.354391 | 28.420479 | 4632 | 18 |
| P13 | 西藏墨竹工卡Mozhugongka, Tibet | 91.468976 | 29.483856 | 3861 | 29 |
| P14 | 西藏工布江达Gongbujiangda, Tibet | 92.311319 | 29.526900 | 4212 | 25 |
| P15 | 西藏林芝Linzhi, Tibet | 94.315592 | 29.337848 | 3195 | 28 |
| P16 | 青海玛沁Maqin, Qinghai | 100.071899 | 34.514712 | 4077 | 31 |
| P17 | 四川若尔盖Ruoergai, Sichuan | 102.949780 | 33.831093 | 3463 | 30 |
| P18 | 四川松潘Songpan, Sichuan | 103.677603 | 32.744826 | 3689 | 30 |
| P19 | 四川康定Kangding, Sichuan | 101.674269 | 30.086797 | 3681 | 30 |
| P20 | 四川理塘Litang, Sichuan | 100.407019 | 30.069332 | 4080 | 30 |
| P21 | 四川炉霍Luhuo, Sichuan | 100.715031 | 31.453404 | 3205 | 30 |
| P22 | 西藏南木林Nanmulin, Tibet | 89.098847 | 29.454476 | 3868 | 19 |
| P23 | 西藏拉孜Lazi, Tibet | 88.050680 | 29.159374 | 4169 | 30 |
| P24 | 青海称多Chengduo, Qinghai | 97.231267 | 33.529197 | 4321 | 25 |
| P25 | 青海囊谦Nangqian, Qinghai | 96.598172 | 31.927747 | 4083 | 30 |
| P26 | 青海久治Jiuzhi, Qinghai | 101.430119 | 33.409655 | 3677 | 30 |
| P27 | 青海都兰Dulan, Qinghai | 97.797869 | 36.062486 | 3003 | 20 |
| P28 | 青海门源Menyuan, Qinghai | 101.418701 | 37.643461 | 3398 | 30 |
| P29 | 青海祁连Qilian, Qinghai | 100.192005 | 38.110367 | 3238 | 13 |
| P30 | 西藏申扎Shenzha, Tibet | 88.687078 | 31.066350 | 4811 | 11 |
表1 蕨麻居群采集信息
Table 1 Populations sampling information of P. anserina
编号 Code | 采集地 Locality | 经度 Longitude (°, E) | 纬度 Latitude (°, N) | 海拔 Altitude (m) | 采集数 Collection number (No.) |
|---|---|---|---|---|---|
| P1 | 青海化隆Hualong, Qinghai | 102.070718 | 36.178380 | 2696 | 22 |
| P2 | 青海泽库Zeku, Qinghai | 101.480690 | 35.012647 | 3625 | 28 |
| P3 | 甘肃碌曲Luqu, Gansu | 102.763742 | 34.818995 | 2972 | 24 |
| P4 | 西藏察雅Chaya, Tibet | 97.057961 | 30.661889 | 4366 | 15 |
| P5 | 西藏八宿Basu, Tibet | 96.791225 | 29.469206 | 3913 | 11 |
| P6 | 西藏左贡Zuogong, Tibet | 97.915733 | 29.674744 | 3997 | 17 |
| P7 | 西藏芒康Mangkang, Tibet | 98.585117 | 29.600331 | 3812 | 25 |
| P8 | 云南香格里拉Shangri-La, Yunnan | 99.666918 | 27.872883 | 3267 | 20 |
| P9 | 西藏安多Anduo, Tibet | 91.514772 | 31.407749 | 4598 | 25 |
| P10 | 西藏比如Biru, Tibet | 93.417949 | 31.286509 | 3909 | 26 |
| P11 | 西藏浪卡子Langkazi, Tibet | 90.237586 | 28.594107 | 4443 | 28 |
| P12 | 西藏措美Cuomei, Tibet | 91.354391 | 28.420479 | 4632 | 18 |
| P13 | 西藏墨竹工卡Mozhugongka, Tibet | 91.468976 | 29.483856 | 3861 | 29 |
| P14 | 西藏工布江达Gongbujiangda, Tibet | 92.311319 | 29.526900 | 4212 | 25 |
| P15 | 西藏林芝Linzhi, Tibet | 94.315592 | 29.337848 | 3195 | 28 |
| P16 | 青海玛沁Maqin, Qinghai | 100.071899 | 34.514712 | 4077 | 31 |
| P17 | 四川若尔盖Ruoergai, Sichuan | 102.949780 | 33.831093 | 3463 | 30 |
| P18 | 四川松潘Songpan, Sichuan | 103.677603 | 32.744826 | 3689 | 30 |
| P19 | 四川康定Kangding, Sichuan | 101.674269 | 30.086797 | 3681 | 30 |
| P20 | 四川理塘Litang, Sichuan | 100.407019 | 30.069332 | 4080 | 30 |
| P21 | 四川炉霍Luhuo, Sichuan | 100.715031 | 31.453404 | 3205 | 30 |
| P22 | 西藏南木林Nanmulin, Tibet | 89.098847 | 29.454476 | 3868 | 19 |
| P23 | 西藏拉孜Lazi, Tibet | 88.050680 | 29.159374 | 4169 | 30 |
| P24 | 青海称多Chengduo, Qinghai | 97.231267 | 33.529197 | 4321 | 25 |
| P25 | 青海囊谦Nangqian, Qinghai | 96.598172 | 31.927747 | 4083 | 30 |
| P26 | 青海久治Jiuzhi, Qinghai | 101.430119 | 33.409655 | 3677 | 30 |
| P27 | 青海都兰Dulan, Qinghai | 97.797869 | 36.062486 | 3003 | 20 |
| P28 | 青海门源Menyuan, Qinghai | 101.418701 | 37.643461 | 3398 | 30 |
| P29 | 青海祁连Qilian, Qinghai | 100.192005 | 38.110367 | 3238 | 13 |
| P30 | 西藏申扎Shenzha, Tibet | 88.687078 | 31.066350 | 4811 | 11 |
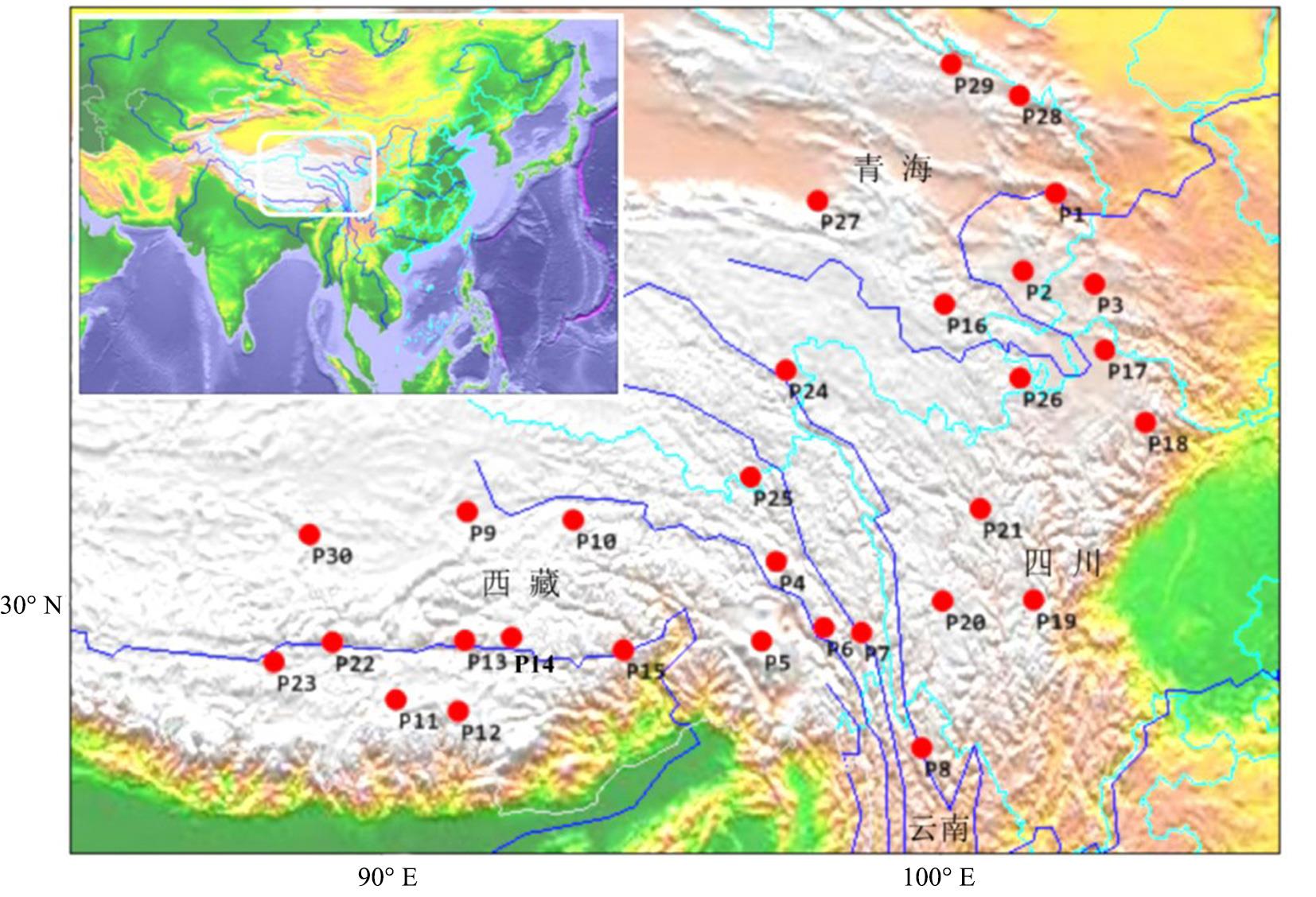
图1 蕨麻采样居群地理分布基于自然资源部标准地图服务网站GS(2019)1822号的标准地图制作,底图边界无修改。Produced based on standard maps from the Department of Natural Resources Standard Map Service website, GS(2019)1822, with no modifications to the base map boundaries.
Fig.1 Geographical distribution of sampling population of P. anserina
片段名称 Name of fragments | 引物名称 Primers name | 引物序列 Sequences of primers (5'-3') | PCR 反应程序 PCR reaction procedures |
|---|---|---|---|
| ITS | 18S | AGGAGAAGT CGTAACAAG | 94 ℃预变性10 min;94 ℃变性1 min、52 ℃退火50 s、72 ℃延伸1 min,30个循环;72 ℃延伸10 min。Pre-denaturation at 94 ℃ for 10 min; Denaturation at 94 ℃ for 1 min, annealing at 52 ℃ for 50 s, reach at 72 ℃ for 1 min, 30 circulates; Reach at 72 ℃ for 10 min. |
| 26A | GTTTCTTTT CCTCCGCT | ||
| trnL-trnF | trnL | AAGAAAAGGC GGAACGACCT | 94 ℃预变性10 min;94 ℃变性1 min、52 ℃退火50 s、72 ℃延伸1 min,32个循环;72 ℃延伸10 min。Pre-denaturation at 94 ℃ for 10 min; Denaturation at 94 ℃ for 1 min, annealing at 52 ℃ for 50 s, reach at 72 ℃ for 1 min, 32 circulates; Reach at 72 ℃ for 10 min. |
| trnF | TCGACCAATTAA ATAATGTGCCAG |
表2 PCR引物信息及反应程序
Table 2 PCR primer information and reaction procedure
片段名称 Name of fragments | 引物名称 Primers name | 引物序列 Sequences of primers (5'-3') | PCR 反应程序 PCR reaction procedures |
|---|---|---|---|
| ITS | 18S | AGGAGAAGT CGTAACAAG | 94 ℃预变性10 min;94 ℃变性1 min、52 ℃退火50 s、72 ℃延伸1 min,30个循环;72 ℃延伸10 min。Pre-denaturation at 94 ℃ for 10 min; Denaturation at 94 ℃ for 1 min, annealing at 52 ℃ for 50 s, reach at 72 ℃ for 1 min, 30 circulates; Reach at 72 ℃ for 10 min. |
| 26A | GTTTCTTTT CCTCCGCT | ||
| trnL-trnF | trnL | AAGAAAAGGC GGAACGACCT | 94 ℃预变性10 min;94 ℃变性1 min、52 ℃退火50 s、72 ℃延伸1 min,32个循环;72 ℃延伸10 min。Pre-denaturation at 94 ℃ for 10 min; Denaturation at 94 ℃ for 1 min, annealing at 52 ℃ for 50 s, reach at 72 ℃ for 1 min, 32 circulates; Reach at 72 ℃ for 10 min. |
| trnF | TCGACCAATTAA ATAATGTGCCAG |

图2 蕨麻cpDNA trnL-trnF片段40种单倍型序列比对的变异位点
Fig.2 Variable sites of the aligned sequences among 40 haplotypes of the cpDNA trnL-trnF fragments of P. anserine (5′-3′)
居群 Population | 个体数 Number of individuals | 遗传多样性 Genetic diversity (h) | 核苷酸多样性Nucleotide diversity (π) | 单倍型种类 Haplotype species (Hd) | 单倍型组成(个体数) Haplotype composition (Number of individuals) |
|---|---|---|---|---|---|
| P1 | 17 | 0.7647 | 0.019448 | 7 | M1(2);M2(1);M3(8);M4(3);M5(1);M6(1);M7(1) |
| P2 | 30 | 0.6828 | 0.013674 | 4 | M1(12);M3(12);M4(3);M7(3) |
| P3 | 17 | 0.6250 | 0.002262 | 4 | M1(9);M4(1);M8(1);M9(6) |
| P4 | 11 | 0.8182 | 0.002797 | 5 | M1(3);M4(2);M5(1);M9(4);M10(1) |
| P5 | 37 | 0.4399 | 0.000702 | 4 | M1(7);M4(27);M5(1);M8(2) |
| P6 | 16 | 0.8333 | 0.024964 | 5 | M1(3);M4(2);M6(3);M11(5);M12(3) |
| P7 | 20 | 0.7000 | 0.034853 | 3 | M1(7);M3(7);M11(6) |
| P8 | 16 | 0.4500 | 0.018146 | 5 | M1(12);M3(1);M6(1);M11(1);M13(1) |
| P9 | 25 | 0.7267 | 0.013210 | 7 | M1(12);M3(4);M4(5);M7(1);M11(1);M14(1);M15(1) |
| P10 | 22 | 0.4372 | 0.002765 | 3 | M1(5);M3(1);M4(16) |
| P11 | 20 | 0.7053 | 0.014960 | 6 | M3(5);M4(2);M5(10);M16(1);M17(1);M18(1) |
| P12 | 18 | 0.8497 | 0.025551 | 10 | M1(2);M3(7);M4(1);M5(1);M8(2);M16(1);M19(1);M20(1);M21(1);M22(1) |
| P13 | 24 | 0.6739 | 0.014432 | 5 | M3(12);M4(7);M20(1);M23(3);M24(1) |
| P14 | 22 | 0.7446 | 0.013573 | 6 | M1(2);M3(2);M4(9);M8(1);M23(7);M25(1) |
| P15 | 20 | 0.6895 | 0.010815 | 7 | M1(1);M3(11);M4(1);M8(3);M23(2);M26(1);M27(1) |
| P16 | 24 | 0.6304 | 0.006351 | 4 | M1(7);M3(3);M4(13);M5(1) |
| P17 | 27 | 0.3960 | 0.005912 | 5 | M1(21);M3(2);M4(2);M28(1);M29(1) |
| P18 | 24 | 0.6123 | 0.008381 | 4 | M1(5);M3(4);M25(14);M30(1) |
| P19 | 17 | 0.5074 | 0.001757 | 5 | M1(12);M4(1);M25(2);M31(1);M32(1) |
| P20 | 27 | 0.7037 | 0.011067 | 6 | M1(14);M3(3);M4(4);M30(2);M33(3);M34(1) |
| P21 | 14 | 0.8132 | 0.014235 | 6 | M1(2);M3(4);M4(5);M30(1);M33(1);M35(1) |
| P22 | 12 | 0.6667 | 0.017032 | 5 | M3(7);M7(1);M20(1);M38(1);M39(2) |
| P23 | 18 | 0.7255 | 0.007721 | 4 | M3(7);M7(6);M20(1);M33(4) |
| P24 | 18 | 0.4641 | 0.000746 | 3 | M1(3);M4(13);M5(2) |
| P25 | 18 | 0.5294 | 0.000870 | 4 | M1(4);M4(12);M25(1);M40(1) |
| P26 | 28 | 0.5000 | 0.011802 | 3 | M1(18);M3(9);M4(1) |
| P27 | 18 | 0.4510 | 0.000710 | 3 | M6(13);M11(4);M12(1) |
| P28 | 26 | 0.6769 | 0.031005 | 5 | M1(12);M3(3);M4(1);M6(1);M11(9) |
| P29 | 9 | 0.9444 | 0.035906 | 7 | M1(2);M3(2);M4(1);M6(1);M11(1);M12(1);M36(1) |
| P30 | 8 | 0.4286 | 0.002536 | 2 | M4(2);M37(6) |
| 总计Overall | 603 | 0.8217 | 0.010641 |
表3 蕨麻cpDNA trnL-trnF片段30个居群的单倍型组成、基因和核苷酸多样性
Table 3 Haplotypes composition, genetic diversity (h) and nucleotide diversity (π) of the trnL-trnF fragment of P. anserina in the 30 populations
居群 Population | 个体数 Number of individuals | 遗传多样性 Genetic diversity (h) | 核苷酸多样性Nucleotide diversity (π) | 单倍型种类 Haplotype species (Hd) | 单倍型组成(个体数) Haplotype composition (Number of individuals) |
|---|---|---|---|---|---|
| P1 | 17 | 0.7647 | 0.019448 | 7 | M1(2);M2(1);M3(8);M4(3);M5(1);M6(1);M7(1) |
| P2 | 30 | 0.6828 | 0.013674 | 4 | M1(12);M3(12);M4(3);M7(3) |
| P3 | 17 | 0.6250 | 0.002262 | 4 | M1(9);M4(1);M8(1);M9(6) |
| P4 | 11 | 0.8182 | 0.002797 | 5 | M1(3);M4(2);M5(1);M9(4);M10(1) |
| P5 | 37 | 0.4399 | 0.000702 | 4 | M1(7);M4(27);M5(1);M8(2) |
| P6 | 16 | 0.8333 | 0.024964 | 5 | M1(3);M4(2);M6(3);M11(5);M12(3) |
| P7 | 20 | 0.7000 | 0.034853 | 3 | M1(7);M3(7);M11(6) |
| P8 | 16 | 0.4500 | 0.018146 | 5 | M1(12);M3(1);M6(1);M11(1);M13(1) |
| P9 | 25 | 0.7267 | 0.013210 | 7 | M1(12);M3(4);M4(5);M7(1);M11(1);M14(1);M15(1) |
| P10 | 22 | 0.4372 | 0.002765 | 3 | M1(5);M3(1);M4(16) |
| P11 | 20 | 0.7053 | 0.014960 | 6 | M3(5);M4(2);M5(10);M16(1);M17(1);M18(1) |
| P12 | 18 | 0.8497 | 0.025551 | 10 | M1(2);M3(7);M4(1);M5(1);M8(2);M16(1);M19(1);M20(1);M21(1);M22(1) |
| P13 | 24 | 0.6739 | 0.014432 | 5 | M3(12);M4(7);M20(1);M23(3);M24(1) |
| P14 | 22 | 0.7446 | 0.013573 | 6 | M1(2);M3(2);M4(9);M8(1);M23(7);M25(1) |
| P15 | 20 | 0.6895 | 0.010815 | 7 | M1(1);M3(11);M4(1);M8(3);M23(2);M26(1);M27(1) |
| P16 | 24 | 0.6304 | 0.006351 | 4 | M1(7);M3(3);M4(13);M5(1) |
| P17 | 27 | 0.3960 | 0.005912 | 5 | M1(21);M3(2);M4(2);M28(1);M29(1) |
| P18 | 24 | 0.6123 | 0.008381 | 4 | M1(5);M3(4);M25(14);M30(1) |
| P19 | 17 | 0.5074 | 0.001757 | 5 | M1(12);M4(1);M25(2);M31(1);M32(1) |
| P20 | 27 | 0.7037 | 0.011067 | 6 | M1(14);M3(3);M4(4);M30(2);M33(3);M34(1) |
| P21 | 14 | 0.8132 | 0.014235 | 6 | M1(2);M3(4);M4(5);M30(1);M33(1);M35(1) |
| P22 | 12 | 0.6667 | 0.017032 | 5 | M3(7);M7(1);M20(1);M38(1);M39(2) |
| P23 | 18 | 0.7255 | 0.007721 | 4 | M3(7);M7(6);M20(1);M33(4) |
| P24 | 18 | 0.4641 | 0.000746 | 3 | M1(3);M4(13);M5(2) |
| P25 | 18 | 0.5294 | 0.000870 | 4 | M1(4);M4(12);M25(1);M40(1) |
| P26 | 28 | 0.5000 | 0.011802 | 3 | M1(18);M3(9);M4(1) |
| P27 | 18 | 0.4510 | 0.000710 | 3 | M6(13);M11(4);M12(1) |
| P28 | 26 | 0.6769 | 0.031005 | 5 | M1(12);M3(3);M4(1);M6(1);M11(9) |
| P29 | 9 | 0.9444 | 0.035906 | 7 | M1(2);M3(2);M4(1);M6(1);M11(1);M12(1);M36(1) |
| P30 | 8 | 0.4286 | 0.002536 | 2 | M4(2);M37(6) |
| 总计Overall | 603 | 0.8217 | 0.010641 |
居群 Population | 个体数 Number of individuals | 遗传多样性 Genetic diversity (h) | 核苷酸多样性Nucleotide diversity (π) | 单倍型种类 Haplotype species (Hd) | 单倍型组成(个体数) Haplotype composition (Number of individuals) |
|---|---|---|---|---|---|
| P1 | 34 | 0.7380 | 0.002867 | 8 | H1(12);H2(13);H3(2);H4(1);H5(2);H6(2);H7(1);H8(1) |
| P2 | 58 | 0.7550 | 0.004208 | 10 | H7(1);H9(5);H10(23);H11(16);H12(7);H13(1);H14(1);H15(1);H16(1);H17(2) |
| P3 | 46 | 0.8918 | 0.006707 | 17 | H2(1);H3(2);H9(2);H10(12);H11(8);H12(5);H15(2);H17(3);H18(1);H19(2); H20(2);H21(1);H22(1);H23(1);H24(1);H25(1);H26(1) |
| P4 | 24 | 0.7319 | 0.002501 | 6 | H9(1);H10(9);H11(9);H12(1);H19(2);H20(2) |
| P5 | 78 | 0.8591 | 0.003594 | 20 | H9(8);H10(21);H11(18);H12(2);H15(2);H17(1);H27(2);H28(1);H29(2);H30(1); H31(7);H32(1);H33(1);H34(2);H35(4);H36(1);H37(1);H38(1);H39(1);H40(1) |
| P6 | 30 | 0.7816 | 0.003028 | 7 | H10(5);H11(11);H12(8);H15(2);H41(1);H42(2);H43(1) |
| P7 | 46 | 0.7227 | 0.002434 | 6 | H9(3);H10(17);H11(16);H12(8);H44(1);H45(1) |
| P8 | 16 | 0.5417 | 0.001108 | 3 | H46(10);H47(5);H48(1) |
| P9 | 48 | 0.7261 | 0.001797 | 8 | H9(15);H10(20);H11(3);H12(5);H42(1);H49(2);H50(1);H51(1) |
| P10 | 50 | 0.6196 | 0.001623 | 9 | H9(6);H10(30);H11(5);H12(1);H15(4);H19(1);H20(1);H50(1);H52(1) |
| P11 | 52 | 0.7278 | 0.002189 | 8 | H9(10);H10(21);H11(15);H19(1);H20(1);H53(2);H54(1);H55(1) |
| P12 | 36 | 0.7317 | 0.001983 | 6 | H9(9);H10(13);H11(11);H19(1);H20(1);H56(1) |
| P13 | 54 | 0.7261 | 0.002297 | 8 | H9(14);H10(21);H11(14);H25(1);H56(1);H57(1);H58(1);H59(1) |
| P14 | 48 | 0.7181 | 0.001760 | 7 | H9(18);H10(16);H11(10);H29(1);H53(1);H60(1);H61(1) |
| P15 | 46 | 0.7034 | 0.002335 | 5 | H9(12);H10(17);H11(15);H62(1);H63(1) |
| P16 | 44 | 0.8161 | 0.005701 | 17 | H1(1);H4(1);H9(2);H10(18);H11(6);H12(3);H14(1);H15(2);H17(1);H19(1); H25(1);H43(1);H61(2);H67(1);H79(1);H102(1);H128(1) |
| P17 | 56 | 0.6818 | 0.001985 | 5 | H9(5);H10(25);H11(19);H12(4);H15(3) |
| P18 | 56 | 0.8344 | 0.007679 | 17 | H1(3);H2(1);H3(1);H9(5);H10(20);H11(10);H12(1);H14(3);H15(1);H25(1); H61(1);H64(1);H65(1);H66(2);H67(3);H68(1);H69(1) |
| P19 | 46 | 0.7072 | 0.002314 | 9 | H9(3);H10(22);H11(12);H12(2);H15(1);H61(2);H70(1);H71(1);H72(2) |
| P20 | 54 | 0.7959 | 0.003749 | 15 | H9(2);H10(20);H11(14);H12(1);H15(1);H19(3);H20(4);H29(2);H30(1);H47(1); H52(1);H73(1);H74(1);H75(1);H76(1) |
| P21 | 156 | 0.8078 | 0.003330 | 21 | H3(1);H9(20);H10(51);H11(34);H12(24);H15(2);H25(1);H49(1);H61(3); H72(6);H76(1);H77(1);H78(1);H79(1);H80(1);H81(1);H82(1);H83(3);H84(1);H85(1);H86(1) |
| P22 | 32 | 0.7359 | 0.002034 | 5 | H9(9);H10(12);H11(8);H125(2);H126(1) |
| P23 | 32 | 0.6290 | 0.001344 | 5 | H9(13);H10(15);H11(2);H29(1);H53(1) |
| P24 | 48 | 0.7660 | 0.002506 | 6 | H9(5);H10(11);H11(13);H12(16);H15(2);H127(1) |
| P25 | 22 | 0.7143 | 0.002165 | 5 | H9(1);H10(10);H11(6);H12(4);H15(1) |
| P26 | 52 | 0.7677 | 0.002383 | 6 | H9(4);H10(17);H11(15);H12(11);H15(4);H43(1) |
| P27 | 66 | 0.8601 | 0.004209 | 21 | H1(6);H2(18);H3(1);H5(1);H11(1);H12(1);H14(2);H87(16);H88(3);H89(1);H90(2);H91(1);H92(2);H93(1);H94(1);H95(1);H96(1);H97(4);H98(1);H99(1);H100(1) |
| P28 | 56 | 0.9565 | 0.012402 | 34 | H1(1);H2(3);H3(1);H4(1);H7(3);H8(2);H9(1);H10(8);H11(8);H12(3); H16(1);H19(1);H20(1);H24(1);H25(1);H29(1);H33(1);H66(1);H67(1);H92(1);H101(1);H102(1);H103(2);H104(1);H105(1);H106(1);H107(1);H108(1);H109(1); H110(1);H111(1);H112(1);H113(1);H114(1) |
| P29 | 24 | 0.9819 | 0.012142 | 19 | H1(2);H3(2);H6(1);H12(1);H13(1);H14(2);H15(1);H24(2);H65(1);H87(2); H92(1);H99(1);H115(1);H116(1);H117(1);H118(1);H119(1);H120(1);H121(1) |
| P30 | 18 | 0.8693 | 0.003152 | 8 | H9(4);H10(2);H11(1);H12(3);H72(5);H122(1);H123(1);H124(1) |
总计 Overall | 1428 | 0.8168 | 0.003584 |
表4 蕨麻ITS片段30个居群的单倍型组成、基因和核苷酸多样性
Table 4 Haplotypes composition, genetic diversity (h) and nucleotide diversity (π) of the ITS fragments of P. anserina in the 30 populations
居群 Population | 个体数 Number of individuals | 遗传多样性 Genetic diversity (h) | 核苷酸多样性Nucleotide diversity (π) | 单倍型种类 Haplotype species (Hd) | 单倍型组成(个体数) Haplotype composition (Number of individuals) |
|---|---|---|---|---|---|
| P1 | 34 | 0.7380 | 0.002867 | 8 | H1(12);H2(13);H3(2);H4(1);H5(2);H6(2);H7(1);H8(1) |
| P2 | 58 | 0.7550 | 0.004208 | 10 | H7(1);H9(5);H10(23);H11(16);H12(7);H13(1);H14(1);H15(1);H16(1);H17(2) |
| P3 | 46 | 0.8918 | 0.006707 | 17 | H2(1);H3(2);H9(2);H10(12);H11(8);H12(5);H15(2);H17(3);H18(1);H19(2); H20(2);H21(1);H22(1);H23(1);H24(1);H25(1);H26(1) |
| P4 | 24 | 0.7319 | 0.002501 | 6 | H9(1);H10(9);H11(9);H12(1);H19(2);H20(2) |
| P5 | 78 | 0.8591 | 0.003594 | 20 | H9(8);H10(21);H11(18);H12(2);H15(2);H17(1);H27(2);H28(1);H29(2);H30(1); H31(7);H32(1);H33(1);H34(2);H35(4);H36(1);H37(1);H38(1);H39(1);H40(1) |
| P6 | 30 | 0.7816 | 0.003028 | 7 | H10(5);H11(11);H12(8);H15(2);H41(1);H42(2);H43(1) |
| P7 | 46 | 0.7227 | 0.002434 | 6 | H9(3);H10(17);H11(16);H12(8);H44(1);H45(1) |
| P8 | 16 | 0.5417 | 0.001108 | 3 | H46(10);H47(5);H48(1) |
| P9 | 48 | 0.7261 | 0.001797 | 8 | H9(15);H10(20);H11(3);H12(5);H42(1);H49(2);H50(1);H51(1) |
| P10 | 50 | 0.6196 | 0.001623 | 9 | H9(6);H10(30);H11(5);H12(1);H15(4);H19(1);H20(1);H50(1);H52(1) |
| P11 | 52 | 0.7278 | 0.002189 | 8 | H9(10);H10(21);H11(15);H19(1);H20(1);H53(2);H54(1);H55(1) |
| P12 | 36 | 0.7317 | 0.001983 | 6 | H9(9);H10(13);H11(11);H19(1);H20(1);H56(1) |
| P13 | 54 | 0.7261 | 0.002297 | 8 | H9(14);H10(21);H11(14);H25(1);H56(1);H57(1);H58(1);H59(1) |
| P14 | 48 | 0.7181 | 0.001760 | 7 | H9(18);H10(16);H11(10);H29(1);H53(1);H60(1);H61(1) |
| P15 | 46 | 0.7034 | 0.002335 | 5 | H9(12);H10(17);H11(15);H62(1);H63(1) |
| P16 | 44 | 0.8161 | 0.005701 | 17 | H1(1);H4(1);H9(2);H10(18);H11(6);H12(3);H14(1);H15(2);H17(1);H19(1); H25(1);H43(1);H61(2);H67(1);H79(1);H102(1);H128(1) |
| P17 | 56 | 0.6818 | 0.001985 | 5 | H9(5);H10(25);H11(19);H12(4);H15(3) |
| P18 | 56 | 0.8344 | 0.007679 | 17 | H1(3);H2(1);H3(1);H9(5);H10(20);H11(10);H12(1);H14(3);H15(1);H25(1); H61(1);H64(1);H65(1);H66(2);H67(3);H68(1);H69(1) |
| P19 | 46 | 0.7072 | 0.002314 | 9 | H9(3);H10(22);H11(12);H12(2);H15(1);H61(2);H70(1);H71(1);H72(2) |
| P20 | 54 | 0.7959 | 0.003749 | 15 | H9(2);H10(20);H11(14);H12(1);H15(1);H19(3);H20(4);H29(2);H30(1);H47(1); H52(1);H73(1);H74(1);H75(1);H76(1) |
| P21 | 156 | 0.8078 | 0.003330 | 21 | H3(1);H9(20);H10(51);H11(34);H12(24);H15(2);H25(1);H49(1);H61(3); H72(6);H76(1);H77(1);H78(1);H79(1);H80(1);H81(1);H82(1);H83(3);H84(1);H85(1);H86(1) |
| P22 | 32 | 0.7359 | 0.002034 | 5 | H9(9);H10(12);H11(8);H125(2);H126(1) |
| P23 | 32 | 0.6290 | 0.001344 | 5 | H9(13);H10(15);H11(2);H29(1);H53(1) |
| P24 | 48 | 0.7660 | 0.002506 | 6 | H9(5);H10(11);H11(13);H12(16);H15(2);H127(1) |
| P25 | 22 | 0.7143 | 0.002165 | 5 | H9(1);H10(10);H11(6);H12(4);H15(1) |
| P26 | 52 | 0.7677 | 0.002383 | 6 | H9(4);H10(17);H11(15);H12(11);H15(4);H43(1) |
| P27 | 66 | 0.8601 | 0.004209 | 21 | H1(6);H2(18);H3(1);H5(1);H11(1);H12(1);H14(2);H87(16);H88(3);H89(1);H90(2);H91(1);H92(2);H93(1);H94(1);H95(1);H96(1);H97(4);H98(1);H99(1);H100(1) |
| P28 | 56 | 0.9565 | 0.012402 | 34 | H1(1);H2(3);H3(1);H4(1);H7(3);H8(2);H9(1);H10(8);H11(8);H12(3); H16(1);H19(1);H20(1);H24(1);H25(1);H29(1);H33(1);H66(1);H67(1);H92(1);H101(1);H102(1);H103(2);H104(1);H105(1);H106(1);H107(1);H108(1);H109(1); H110(1);H111(1);H112(1);H113(1);H114(1) |
| P29 | 24 | 0.9819 | 0.012142 | 19 | H1(2);H3(2);H6(1);H12(1);H13(1);H14(2);H15(1);H24(2);H65(1);H87(2); H92(1);H99(1);H115(1);H116(1);H117(1);H118(1);H119(1);H120(1);H121(1) |
| P30 | 18 | 0.8693 | 0.003152 | 8 | H9(4);H10(2);H11(1);H12(3);H72(5);H122(1);H123(1);H124(1) |
总计 Overall | 1428 | 0.8168 | 0.003584 |
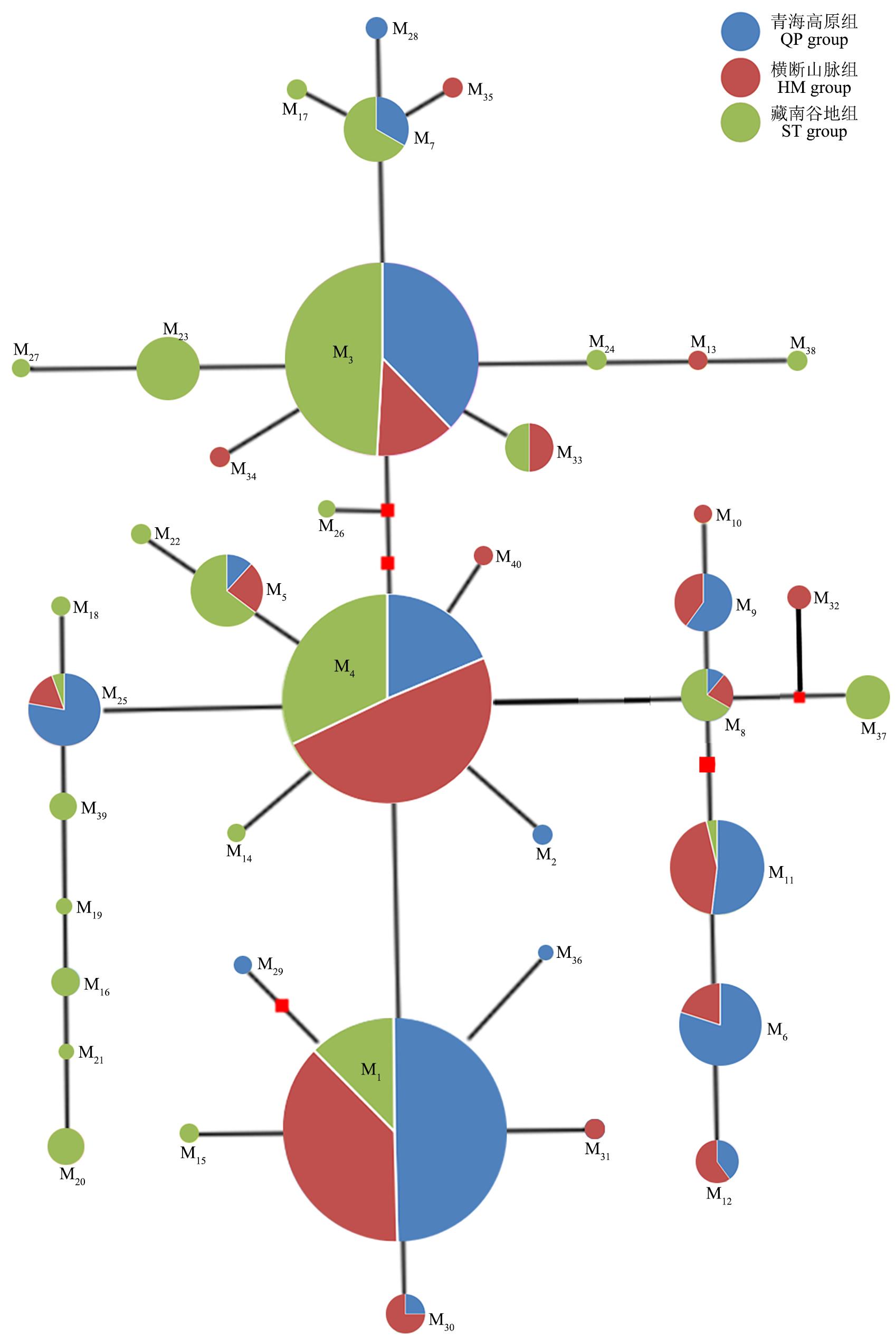
图4 基于不同分组的蕨麻cpDNA trnL-trnF片段中央连接网状图
Fig.4 Median-joining network of the haplotypes detected from the cpDNA trnL-trnF fragments of P. anserina based on different groups
类群 Population groups | cpDNA trnL-trnF | ITS | ||||||
|---|---|---|---|---|---|---|---|---|
HS (P值P-value) | HT (P值P-value) | GST (P值P-value) | NST (P值P-value) | HS (P值P-value) | HT (P值P-value) | GST (P值P-value) | NST (P值P-value) | |
青海高原组 QP group | 0.628 (0.0501) | 0.822 (0.0441) | 0.235 (0.0678) | 0.465 (0.1930)** | 0.823 (0.0296) | 0.907 (0.0334) | 0.092 (0.0284) | 0.227 (0.0250) |
横断山脉组 HM group | 0.626 (0.0518) | 0.793 (0.0367) | 0.211 (0.0395) | 0.358 (0.0706)** | 0.739 (0.0259) | 0.819 (0.0350) | 0.098 (0.0678) | 0.607 (0.2090)** |
藏南谷地组 ST group | 0.665 (0.0419) | 0.867 (0.0218) | 0.234 (0.0594) | 0.307 (0.1057) | 0.719 (0.0213) | 0.748 (0.0300) | 0.038 (0.0133) | 0.050 (P>0.05) |
所有类群 All populations | 0.640 (0.0270) | 0.849 (0.0197) | 0.246 (0.0319) | 0.417 (0.0953)** | 0.763 (0.0172) | 0.844 (0.0245) | 0.096 (0.0258) | 0.522 (0.1020)** |
表5 蕨麻cpDNA trnL-trnF片段和ITS片段遗传多样性分析
Table 5 Genetic diversity analysis of cpDNA trnL-rnF fragments and ITS fragments of P. anserine
类群 Population groups | cpDNA trnL-trnF | ITS | ||||||
|---|---|---|---|---|---|---|---|---|
HS (P值P-value) | HT (P值P-value) | GST (P值P-value) | NST (P值P-value) | HS (P值P-value) | HT (P值P-value) | GST (P值P-value) | NST (P值P-value) | |
青海高原组 QP group | 0.628 (0.0501) | 0.822 (0.0441) | 0.235 (0.0678) | 0.465 (0.1930)** | 0.823 (0.0296) | 0.907 (0.0334) | 0.092 (0.0284) | 0.227 (0.0250) |
横断山脉组 HM group | 0.626 (0.0518) | 0.793 (0.0367) | 0.211 (0.0395) | 0.358 (0.0706)** | 0.739 (0.0259) | 0.819 (0.0350) | 0.098 (0.0678) | 0.607 (0.2090)** |
藏南谷地组 ST group | 0.665 (0.0419) | 0.867 (0.0218) | 0.234 (0.0594) | 0.307 (0.1057) | 0.719 (0.0213) | 0.748 (0.0300) | 0.038 (0.0133) | 0.050 (P>0.05) |
所有类群 All populations | 0.640 (0.0270) | 0.849 (0.0197) | 0.246 (0.0319) | 0.417 (0.0953)** | 0.763 (0.0172) | 0.844 (0.0245) | 0.096 (0.0258) | 0.522 (0.1020)** |

图6 蕨麻cpDNA trnL-trnF片段单倍型地理分布基于自然资源部标准地图服务网站GS(2019)1822号的标准地图制作,底图边界无修改。Produced based on standard maps from the Department of Natural Resources Standard Map Service website, GS(2019)1822, with no modifications to the base map boundaries.
Fig.6 The geographical distribution information of the haplotypes detected from the cpDNA trnL-trnF fragments of P. anserina

图7 蕨麻ITS片段单倍型地理分布饼状图表示每个居群中单倍型的频率,下同。基于自然资源部标准地图服务网站GS(2019)1822号的标准地图制作,底图边界无修改。The pie charts show the proportions of haplotypes within each population, the same below. Produced based on standard maps from the Department of Natural Resources Standard Map Service website, GS(2019)1822, with no modifications to the base map boundaries.
Fig.7 The geographical distribution information of the haplotypes detected from the ITS fragments of P. anserina
组别 Group | 变异来源 Source of variation | cpDNA trnL-trnF | ITS | ||
|---|---|---|---|---|---|
| 变异百分比Percentage of variation (%) | 群体间分化指数 Fixation index (FST) | 变异百分比 Percentage of variation (%) | 群体间分化指数 Fixation index (FST) | ||
总组别 Overall group | 居群间Among populations | 40.31 | 0.40313** | 45.91 | 0.45911** |
| 居群内Within populations | 59.69 | 54.09 | |||
青海高原组 QP group | 居群间Among populations | 46.33 | 0.46333** | 49.74 | 0.49739** |
| 居群内Within populations | 53.67 | 50.26 | |||
横断山脉组 HM group | 居群间Among populations | 35.16 | 0.35160** | 33.36 | 0.33357** |
| 居群内Within populations | 64.84 | 66.64 | |||
藏南谷地组 ST group | 居群间Among populations | 27.08 | 0.27075** | 4.15 | 0.04148** |
| 居群内Within populations | 72.92 | 95.85 | |||
表6 基于cpDNA trnL-trnF和ITS对蕨麻居群的分子变异分析
Table 6 Analyses of molecular variance (AMOVA) based on cpDNA trnL-trnF and ITS dataset from P. anserina
组别 Group | 变异来源 Source of variation | cpDNA trnL-trnF | ITS | ||
|---|---|---|---|---|---|
| 变异百分比Percentage of variation (%) | 群体间分化指数 Fixation index (FST) | 变异百分比 Percentage of variation (%) | 群体间分化指数 Fixation index (FST) | ||
总组别 Overall group | 居群间Among populations | 40.31 | 0.40313** | 45.91 | 0.45911** |
| 居群内Within populations | 59.69 | 54.09 | |||
青海高原组 QP group | 居群间Among populations | 46.33 | 0.46333** | 49.74 | 0.49739** |
| 居群内Within populations | 53.67 | 50.26 | |||
横断山脉组 HM group | 居群间Among populations | 35.16 | 0.35160** | 33.36 | 0.33357** |
| 居群内Within populations | 64.84 | 66.64 | |||
藏南谷地组 ST group | 居群间Among populations | 27.08 | 0.27075** | 4.15 | 0.04148** |
| 居群内Within populations | 72.92 | 95.85 | |||
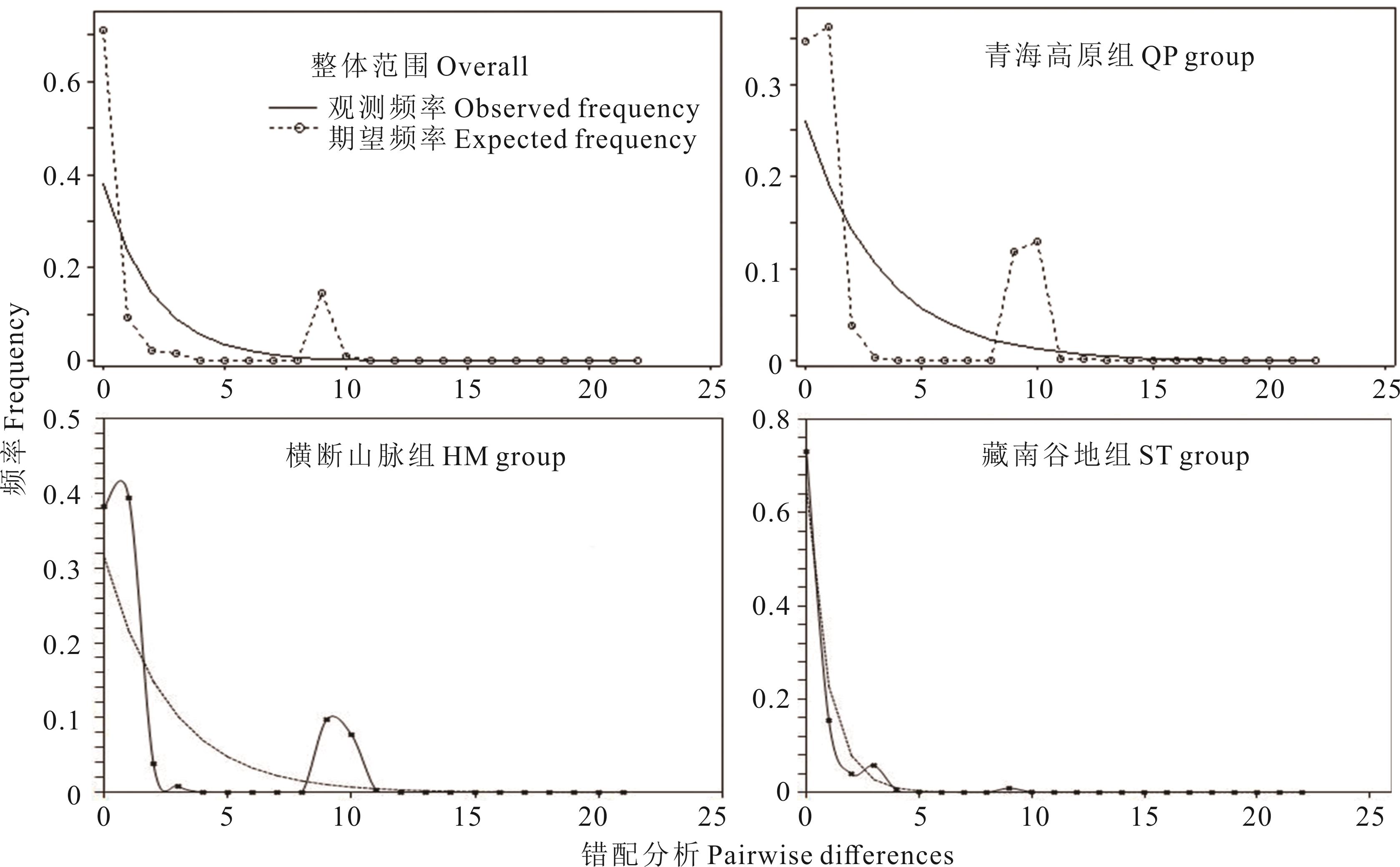
图8 cpDNA trnL-trnF序列所有个体的歧点分布分析Exp: 期望值曲线Expected value curve; Obs: 观测值曲线Observed value curve. 下同The same below.
Fig.8 The mismatch distribution for cpDNA trnL-trnF data of all individuals
组别 Groups | 中性检验Neutrality tests | 错配分布Mismatch distribution | ||||
|---|---|---|---|---|---|---|
Tajima’s D (P值 P-value) | Fu’s FS (P值 P-value) | Fu and Li’s D (P值 P-value) | Fu and Li’s F (P值 P-value) | SSD (P值 P-value) | HRag (P值 P-value) | |
总组别 Overall group | 0.11976 | 5.72410 | -1.14153 | -1.38361 | 0.00926 | 0.02950 |
| (0.56557) | (0.81313) | (P>0.10) | (P>0.10) | (0.03133)* | (0.07433) | |
青海高原组 QP group | 0.54005 | 19.44008 | 0.22999 | 0.40698 | 0.04831 | 0.07662 |
| (0.74500) | (0.98600) | (P>0.10) | (P>0.10) | (0.31100) | (0.50200) | |
横断山脉组 HM group | -0.64632 | 6.90646 | -1.73049 | -1.58040 | 0.03810 | 0.07003 |
| (0.30000) | (0.92400) | (P>0.10) | (P>0.10) | (0.52900) | (0.65600) | |
藏南谷地组 ST group | -2.04300 | 4.00364 | -4.51770 | -4.32336 | 0.04989 | 0.06099 |
| (0.00100)** | (0.86500) | (P<0.02)** | (P<0.02)** | (0.19200) | (0.49400) | |
表7 蕨麻cpDNA trnL-trnF片段中性检验及分子错配分析
Table 7 Neutrality tests and mismatch distribution analysis for cpDNA trnL-trnF fragments of P. anserina
组别 Groups | 中性检验Neutrality tests | 错配分布Mismatch distribution | ||||
|---|---|---|---|---|---|---|
Tajima’s D (P值 P-value) | Fu’s FS (P值 P-value) | Fu and Li’s D (P值 P-value) | Fu and Li’s F (P值 P-value) | SSD (P值 P-value) | HRag (P值 P-value) | |
总组别 Overall group | 0.11976 | 5.72410 | -1.14153 | -1.38361 | 0.00926 | 0.02950 |
| (0.56557) | (0.81313) | (P>0.10) | (P>0.10) | (0.03133)* | (0.07433) | |
青海高原组 QP group | 0.54005 | 19.44008 | 0.22999 | 0.40698 | 0.04831 | 0.07662 |
| (0.74500) | (0.98600) | (P>0.10) | (P>0.10) | (0.31100) | (0.50200) | |
横断山脉组 HM group | -0.64632 | 6.90646 | -1.73049 | -1.58040 | 0.03810 | 0.07003 |
| (0.30000) | (0.92400) | (P>0.10) | (P>0.10) | (0.52900) | (0.65600) | |
藏南谷地组 ST group | -2.04300 | 4.00364 | -4.51770 | -4.32336 | 0.04989 | 0.06099 |
| (0.00100)** | (0.86500) | (P<0.02)** | (P<0.02)** | (0.19200) | (0.49400) | |
组别 Groups | 中性检验Neutrality tests | 错配分布Mismatch distribution | ||||
|---|---|---|---|---|---|---|
Tajima’s D (P值 P-value) | Fu’s FS (P值 P-value) | Fu and Li’s D (P值 P-value) | Fu and Li’s FS (P值 P-value) | SSD (P值 P-value) | HRag (P值 P-value) | |
总组别 Overall group | -1.31350 | -24.78898 | -6.78954 | -5.02878 | 0.01943 | 0.03285 |
| (0.04000)* | (0.00300)** | (P<0.02)** | (P<0.02)** | (0.15900) | (0.44600) | |
青海高原组 QP group | 0.42414 | -24.45324 | -4.56846 | -3.08924 | 0.03085 | 0.02126 |
| (0.66800) | (0.00000)** | (P<0.02)** | (P<0.02)** | (0.20000) | (0.92000) | |
横断山脉组 HM group | -1.93781 | -26.94711 | -3.60133 | -3.27625 | 0.00566 | 0.08246 |
| (0.00100)** | (0.00000)** | (P<0.02)** | (P<0.02)** | (0.00000)** | (0.00000)** | |
藏南谷地组 ST group | -1.93781 | -26.94711 | -2.98723 | -3.09026 | 0.00565 | 0.08246 |
| (0.00000)** | (0.00000)** | (P<0.05)* | (P<0.02)** | (0.00000)** | (0.00000)** | |
表8 蕨麻ITS片段中性检验及分子错配分析
Table 8 Neutrality tests and mismatch distribution analysis for ITS fragments of P. anserina
组别 Groups | 中性检验Neutrality tests | 错配分布Mismatch distribution | ||||
|---|---|---|---|---|---|---|
Tajima’s D (P值 P-value) | Fu’s FS (P值 P-value) | Fu and Li’s D (P值 P-value) | Fu and Li’s FS (P值 P-value) | SSD (P值 P-value) | HRag (P值 P-value) | |
总组别 Overall group | -1.31350 | -24.78898 | -6.78954 | -5.02878 | 0.01943 | 0.03285 |
| (0.04000)* | (0.00300)** | (P<0.02)** | (P<0.02)** | (0.15900) | (0.44600) | |
青海高原组 QP group | 0.42414 | -24.45324 | -4.56846 | -3.08924 | 0.03085 | 0.02126 |
| (0.66800) | (0.00000)** | (P<0.02)** | (P<0.02)** | (0.20000) | (0.92000) | |
横断山脉组 HM group | -1.93781 | -26.94711 | -3.60133 | -3.27625 | 0.00566 | 0.08246 |
| (0.00100)** | (0.00000)** | (P<0.02)** | (P<0.02)** | (0.00000)** | (0.00000)** | |
藏南谷地组 ST group | -1.93781 | -26.94711 | -2.98723 | -3.09026 | 0.00565 | 0.08246 |
| (0.00000)** | (0.00000)** | (P<0.05)* | (P<0.02)** | (0.00000)** | (0.00000)** | |

图10 青藏高原蕨麻气候生态位模型估算底图显示4个时间的估算模型,分别为末次最大间冰期、末次盛冰期、全新世中期以及当前。基于自然资源部标准地图服务网站GS(2019)1822号的标准地图制作,底图边界无修改。Image shows distribution models of the four periods, last interglacial maximum, last glacial maximum, mid-holocene, and current. Produced based on standard maps from the Department of Natural Resources Standard Map Service website, GS(2019)1822, with no modifications to the base map boundaries.
Fig.10 Estimated climatic niche models for P. anserina in Qinghai-Tibet Plateau
| 1 | Qiu Y X, Lu Q X, Zhang Y H, et al. Phylogeography of East Asia’s Tertiary relict plants: Current progress and future prospects. Biodiversity Science, 2017, 25(2): 136-146. |
| 邱英雄, 鹿启祥, 张永华, 等. 东亚第三纪孑遗植物的亲缘地理学: 现状与趋势. 生物多样性, 2017, 25(2): 136-146. | |
| 2 | Spicer R, Harris N, Widdowson M, et al. Constant elevation of southern Tibet over the past 15 million years. Nature, 2003, 421(6923): 622-624. |
| 3 | Renner S S. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. Journal of Biogeography, 2016, 43(8): 1479-1487. |
| 4 | Spicer R A. Tibet, the Himalaya, Asian monsoons and biodiversity-In what ways are they related? Plant Diversity, 2017, 39(5): 233-244. |
| 5 | Davis M B, Shaw R G. Range shifts and adaptive responses to quaternary climate change. Science, 2001, 292(5517): 673-679. |
| 6 | Hewitt G M. Genetic consequences of climatic oscillations in the quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences, 2004, 359(1442): 183-195. |
| 7 | Liu C Z, Xu B, Li Z M, et al. Phytogeographical studies on the endemic genera of seed plants from Hengduan Mountains. Journal of Yunnan Normal University (Natural Sciences Edition), 2012, 32(1): 72-78. |
| 刘常周, 徐波, 李志敏, 等. 横断山区种子植物特有属的植物地理学研究. 云南师范大学学报(自然科学版), 2012, 32(1): 72-78. | |
| 8 | Myers N, Mittermeier R A, Mittermeier C G, et al. Biodiversity hotspots for conservation priorities. Nature, 2000, 403(6772): 853-858. |
| 9 | Xing Y W, Ree R H. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proceedings of the National Academy of Sciences, 2017, 114(17): E3444-E3451. |
| 10 | Zhang Q, Chang T Y, George M, et al. Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Molecular Ecology, 2005, 14(11): 3513-3524. |
| 11 | Sheng Y F, Fei L Y, Xin D, et al. Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and ITS correlation with the quaternary climate change. Molecular Ecology, 2008, 17(23): 5135-5145. |
| 12 | Wang L Y, Abbott R J, Zheng W, et al. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Molecular Ecology, 2009, 18(4): 709-721. |
| 13 | Rui J D, Abbott R J, Liang L T, et al. Out of the Qinghai-Tibet Plateau: Evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophaë rhamnoides (Elaeagnaceae). New Phytologist, 2012, 194(4): 1123-1133. |
| 14 | Zhang H X, Zhang M L. Identifying a contact zone between two phylogeographic lineages of Clematis sibirica (Ranunculeae) in the Tianshan and Altai Mountains. Journal of Systematics & Evolution, 2012, 50(4): 295-304. |
| 15 | Li J, Yan P Y, Qian F J, et al. The Saussurea involucrata transcriptome knowledge base. Chinese Bulletin of Botany, 2017, 52(4): 530-538. |
| 李锦, 严潘瑶, 钱飞箭, 等. 新疆天山雪莲转录组注解知识库. 植物学报, 2017, 52(4): 530-538. | |
| 16 | Yang Y, Chen J G, Song B, et al. Advances in the studies of plant diversity and ecological adaptation in the subnival ecosystem of the Qinghai-Tibet Plateau. Chinese Science Bulletin, 2019, 64(27): 2856-2864. |
| 杨扬, 陈建国, 宋波, 等. 青藏高原冰缘植物多样性与适应机制研究进展. 科学通报, 2019, 64(27): 2856-2864. | |
| 17 | Liu S J, Li S Y, Song J H, et al. Anticancer effects of polysaccharide from Potentilla anserine L. Chinese Journal of Modern Applied Pharmacy, 2011, 28(3): 185-188. |
| 刘素君, 李世元, 宋九华, 等. 鹅绒委陵菜多糖抗肿瘤作用研究. 中国现代应用药学, 2011, 28(3): 185-188. | |
| 18 | Zhang L, Li J Y, Jin X, et al. Possible involvement of alpha B-crystallin in the cardio protective effect of n-butanol extract of Potentilla anserina L. on myocardial ischemia/reperfusion injury in rat. Phytomedicine, 2019, 55: 320-329. |
| 19 | Wu H P, Zhong Q L, Zhang Z, et al. Effect of extract of Potentilla anserina L. on alpha-glucosidase activity. Traditional Chinese Drug Research & Clinical Pharmacology, 2010, 21(1): 4-6. |
| 吴慧平, 仲启亮, 张喆, 等. 蕨麻提取物对α-葡萄糖苷酶活性的影响. 中药新药与临床药理, 2010, 21(1): 4-6. | |
| 20 | Zhao B T, Tao F Q, Wang J L, et al. The sulfated modification and antioxidative activity of polysaccharides from Potentilla anserine L. New Journal of Chemistry, 2020, 44(12): 4726-4735. |
| 21 | Angela M, David L, Lena F, et al. Phytochemical composition of Potentilla anserina L. analyzed by an integrative GC-MS and LC-MS metabolomics platform. Metabolomics, 2013, 9(3): 599-607. |
| 22 | Li J Q, Cai G M, Li L Z. Chinese Juema. Beijing: Science Press, 2020. |
| 李军乔, 蔡光明, 李灵芝. 中国蕨麻. 北京: 科学出版社, 2020. | |
| 23 | Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora reipublicae popularis sinicae (Volume 37). Beijing: Science Press, 1996: 275. |
| 中国科学院中国植物志编辑委员会. 中国植物志(第三十七卷). 北京: 科学出版社, 1996: 275. | |
| 24 | Wu Z Y, Raven P H, Hong D Y. Flora of China (Volume 9): Pittosporaceae through Connaraceae. Missouri: Missouri Botanical Garden Press, 2003. |
| 25 | Fu G, Li J Q, Bao J Y, et al. Development of microsatellite primers in Potentilla anserina by magnetic beads enrichment. Acta Prataculturae Sinica, 2018, 27(2): 124-134. |
| 富贵, 李军乔, 包锦渊, 等. 磁珠富集法开发蕨麻SSR标记引物. 草业学报, 2018, 27(2): 124-134. | |
| 26 | Liu H H. SSR primer development and henetic diversity analysis of Potentilla anserine germplasm resources. Xining: Qinghai Minzu University, 2017. |
| 刘贺贺. 蕨麻SSR引物开发及种质资源遗传多样性分析. 西宁: 青海民族大学, 2017. | |
| 27 | Ma B, Li J Q, Liu H H, et al. Construction and analysis of genetic similarity of SSR fingerprints in Juema. Molecular Plant Breeding, 2019, 17(13): 4367-4377. |
| 马斌, 李军乔, 刘贺贺, 等. 蕨麻品种SSR指纹图谱的构建及遗传相似性分析. 分子植物育种, 2019, 17(13): 4367-4377. | |
| 28 | Doyle J J. A rapid total DNA preparation procedure for fresh plant tissue. Focus, 1990, 12: 13-15. |
| 29 | Favre A, Yuan Y M, Küpfer P, et al. Phylogeny of subtribe Gentianinae (Gentianaceae): Biogeographic inferences despite limitations in temporal calibration points. Taxon, 2010, 59(6): 1701-1711. |
| 30 | He T N, Liu S W, Chen S L. Nomenclatural novelities in Swertia (Gentianaceae). Plant Diversity and Resources, 2013, 35(3): 386-392. |
| 31 | Xu Z Q, Yang J S, Hou Z Q, et al. The progress in the study of continental dynamics of the Tibetan Plateau. Geology in China, 2016, 43(1): 1-42. |
| 许志琴, 杨经绥, 侯增谦, 等. 青藏高原大陆动力学研究若干进展. 中国地质, 2016, 43(1): 1-42. | |
| 32 | Xu Z Q, Yang J S, Li H B, et al. On the tectonics of the India-Asia collision. Acta Geologica Sinica, 2011, 85(1): 1-33. |
| 许志琴, 杨经绥, 李海兵, 等. 印度-亚洲碰撞大地构造. 地质学报, 2011, 85(1): 1-33. | |
| 33 | Gao L Z, Zhang C H, Chang L P, et al. Microsatellite diversity within Oryza sativa with emphasis on indica-japonica divergence. Genetics Research, 2005, 85(1): 1-14. |
| 34 | Zhu B, Kidd W S F, Rowley D B, et al. Age of initiation of the India-Asia collision in the east-central Himalaya. The Journal of Geology, 2005, 113(3): 265-285. |
| [1] | 谭湘蛟, 董逵才, 张华, 唐川川, 杨燕. 积雪增加对青藏高原高寒草甸土壤磷有效性的影响[J]. 草业学报, 2024, 33(7): 205-214. |
| [2] | 田甜, 李军乔, 马斌, 王鑫慈, 曲俊儒. 基于SSR分子标记的自然状态下蕨麻采样策略研究[J]. 草业学报, 2023, 32(9): 181-197. |
| [3] | 张东, 侯晨, 马文明, 王长庭, 邓增卓玛, 张婷. 高寒草地不同灌丛化梯度下土壤酶活性研究[J]. 草业学报, 2023, 32(9): 79-92. |
| [4] | 杨欣怡, 杨富强, 周旭姣, 王明军, 黄海霞, 鲁松松, 张晓玮, 杜伟波, 王旭虎, 田青, 赵安, 贺万鹏, 周晓雷. 青藏高原东北边缘云杉-巴山冷杉林火烧迹地草本植物群落构建机理[J]. 草业学报, 2023, 32(8): 40-47. |
| [5] | 周晓雷, 杨富强, 王明军, 黄海霞, 田青, 周旭姣, 赵安, 贺万鹏, 赵艳丽, 姜礼红. 青藏高原东北边缘云杉-巴山冷杉林火烧迹地草本植物群落主要种生态位特征[J]. 草业学报, 2023, 32(7): 23-37. |
| [6] | 周娟娟, 刘云飞, 王敬龙, 魏巍. 短期养分添加对西藏沼泽化高寒草甸地上生物量、植物多样性和功能性状的影响[J]. 草业学报, 2023, 32(11): 17-29. |
| [7] | 王瑞泾, 冯琦胜, 金哲人, 刘洁, 赵玉婷, 葛静, 梁天刚. 青藏高原退化草地的恢复潜势研究[J]. 草业学报, 2022, 31(6): 11-22. |
| [8] | 李洋, 王毅, 韩国栋, 孙建, 汪亚峰. 青藏高原高寒草地土壤微生物量碳氮含量特征及其控制要素[J]. 草业学报, 2022, 31(6): 50-60. |
| [9] | 蔺豆豆, 琚泽亮, 柴继宽, 赵桂琴. 青藏高原燕麦附着耐低温乳酸菌的筛选与鉴定[J]. 草业学报, 2022, 31(5): 103-114. |
| [10] | 李晨芹, 李军乔, 王鑫慈, 牛永昆, 曲俊儒. 蕨麻根腐病病原菌的分离鉴定及其生物学特性研究[J]. 草业学报, 2022, 31(4): 113-123. |
| [11] | 李颖, 吴静, 李纯斌, 秦格霞. 2003-2018年青藏高原草地的地表层土壤热通量时空变化[J]. 草业学报, 2022, 31(11): 1-14. |
| [12] | 金哲人, 冯琦胜, 王瑞泾, 梁天刚. 基于MODIS数据与机器学习的青藏高原草地地上生物量研究[J]. 草业学报, 2022, 31(10): 1-17. |
| [13] | 付刚, 王俊皓, 李少伟, 何萍. 藏北高寒草地牧草营养品质对放牧的响应机制[J]. 草业学报, 2021, 30(9): 38-50. |
| [14] | 南志标, 王彦荣, 聂斌, 李春杰, 张卫国, 夏超. 春箭筈豌豆新品种“兰箭3号”选育与特性评价[J]. 草业学报, 2021, 30(4): 111-120. |
| [15] | 吴瑞, 刘文辉, 张永超, 秦燕, 魏小星, 刘敏洁. 青藏高原老芒麦落粒性及农艺性状相关性研究[J]. 草业学报, 2021, 30(4): 130-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||